Why Alt-R HDR Enhancer Protein?
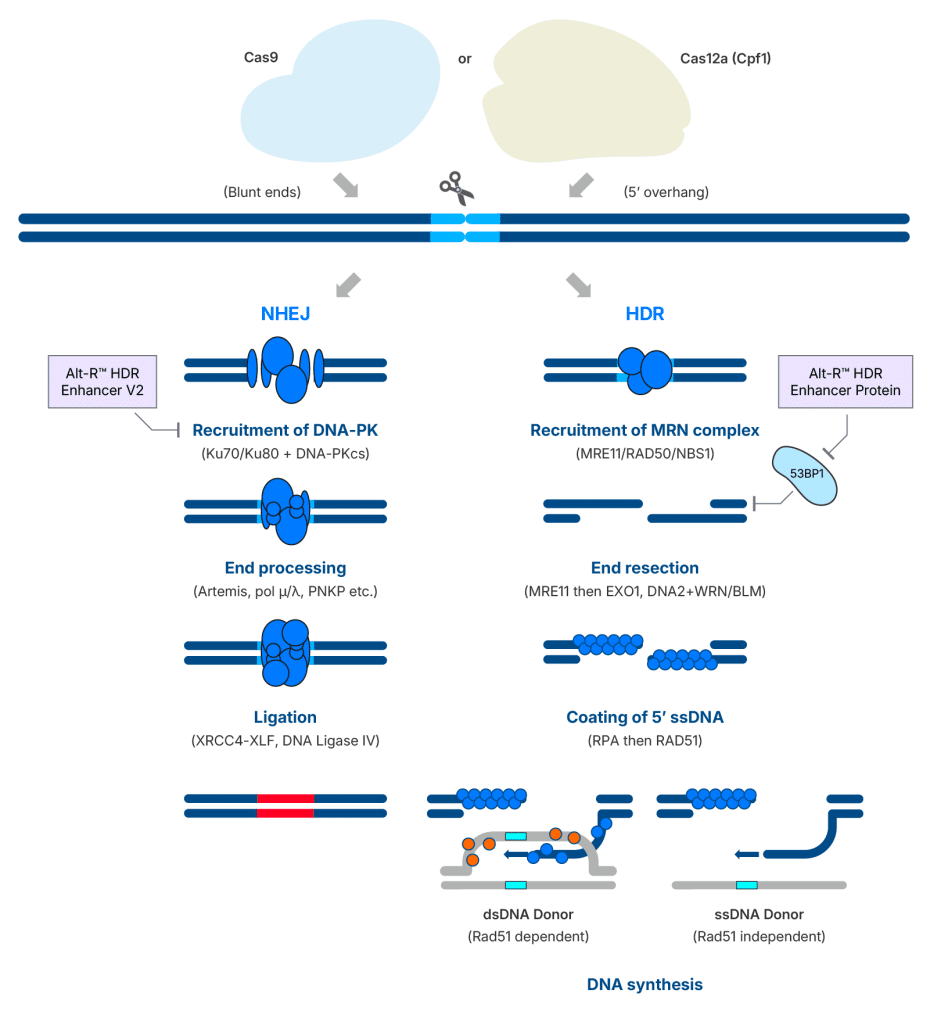
One of the most effective ways to achieve precise genome editing is by inducing a double-strand break (DSB) using a sequence-specific nuclease, such as CRISPR-Cas9, while simultaneously providing a single- or double-stranded DNA template to guide homology-directed repair (HDR). However, cells have a natural preference for repairing DSBs through non-homologous end joining (NHEJ), a fast but error-prone pathway that simply rejoins broken DNA ends. HDR, by contrast, relies on a donor DNA template to make precise edits but occurs far less frequently, and its efficiency is highly dependent on cell type, cell cycle stage, and target site.
A key regulator of this repair pathway choice is the p53-binding protein 1 (53BP1) (Figure 1). This protein is rapidly recruited to DSBs, where it suppresses end resection, a critical step for HDR, thereby tilting the balance in favor of NHEJ. A modified ubiquitin variant known as i53 (inhibitor of 53BP1) has previously been shown to block 53BP1 recruitment and promote HDR when delivered as mRNA or plasmid DNA [1]. However, our team found that i53 effectiveness was highly site dependent when delivered as a purified protein, limiting its potential for streamlined genome editing workflows. To overcome this limitation, we generated a library of ubiquitin variants and conducted a two-hybrid screen to identify variants with improved binding affinity for a Tudor domain containing fragment of 53BP1 that were then verified using Bio-Layer interferometry. Mutations that improved the in vitro affinity were combined to generate HDR Enhancer Protein (HEP). HEP has ~50 fold higher affinity for the Tudor domain of 53BP1 compared to i53. The improved affinity of HEP for 53BP1 relative to i53 translates to improved consistency in boosting HDR when delivered as protein and reduces the dose needed to maximize HDR (Figure 2).
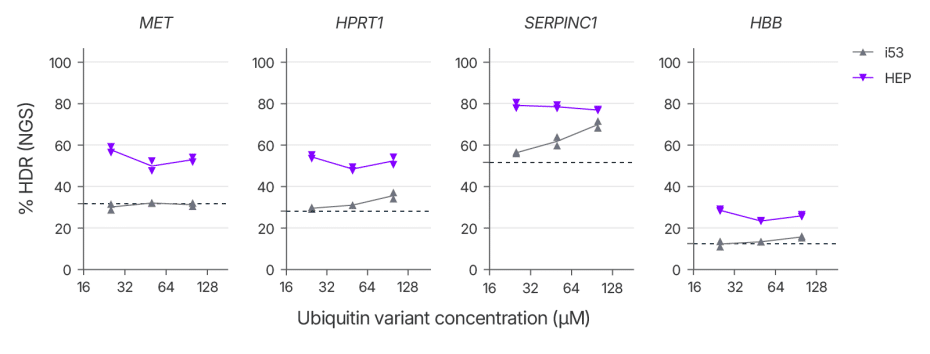
Alt-R HDR Enhancer Protein increases HDR efficiency across multiple loci and cell types
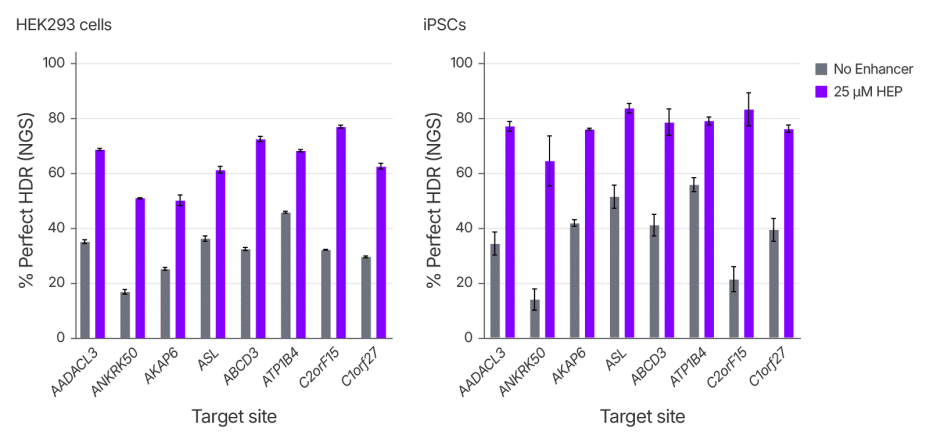
Achieving efficient and precise gene editing, especially in hard-to-edit cell types, remains a significant challenge for CRISPR researchers. To evaluate the performance of Alt-R HDR Enhancer Protein across cell types, we measured rates of insertion of a 6 bp sequence by HDR at eight genomic loci in two different cell lines, the commonly used HEK293 line and primary human induced pluripotent stem cells (iPSCs), a more translationally relevant but difficult-to-edit cell type. Perfect HDR rates increased across all loci, with a median increase of ~2 fold for both cell types. (Figure 3). These results highlight the effectiveness of Alt-R HDR Enhancer in workflows for basic research or translational applications.
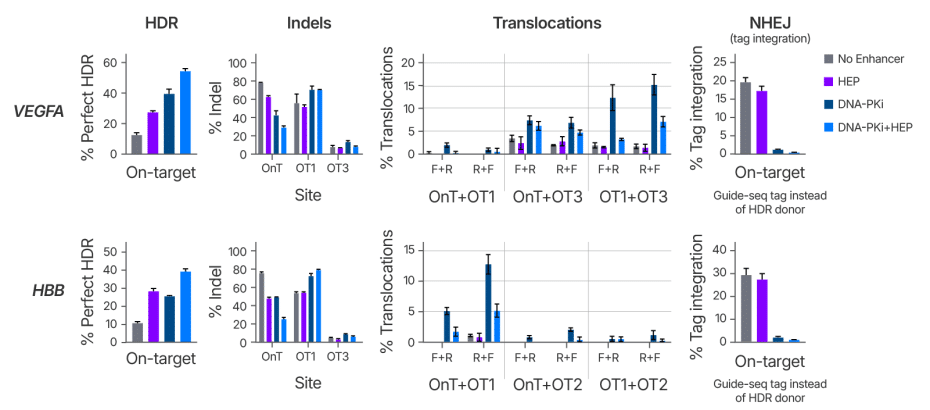
Alt-R HDR Enhancer Protein boosts HDR without increasing the frequency of translocations or off-target indels
A key consideration in genome editing is achieving high on-target editing without introducing unintended mutations. In addition to indel formation, unintended chromosomal rearrangements such as translocations can pose serious risks to genomic integrity. Recent studies have highlighted the risk of NHEJ inhibition for genome stability [2, 3]. While 53BP1 can be thought of as an anti-HDR factor, it is also considered pro-NHEJ as resected DNA ends are resistant to NHEJ [4]. To determine what effect 53BP1 inhibition has on NHEJ repair and overall genotoxicity, we assessed translocations between known cut sites for sgRNAs with highly edited off-target sites. HEK293 cells were treated with Cas9 RNP with and without HEP alongside either a short dsDNA guide-seq tag or ssDNA HDR donor template. Cells were then plated in control media or media containing a small-molecule DNA-PKcs inhibitor (Alt-R HDR Enhancer V2) with editing after 48 hours analyzed by NGS (Figure 4). Both HEP and Alt-R HDR Enhancer V2 increased rates of HDR and use of HEP and V2 together further increased HDR rates, consistent with them working through distinct mechanisms. While HEP had minimal effects on off-target indel formation, V2 increased off-target indels, likely due to a shift from potentially perfect repair of breaks by NHEJ to inherently mutagenic MMEJ repair. Consistent with recent studies, we found that that inhibition of DNA-PKcs increased translocation frequency as quantified by PASTA (Primer Anchored Statistical Translocation Analysis) [5], however no increase in translocations was observed with HEP treatment. To better understand how HEP is affecting DNA repair, we looked at the cells treated with guide-seq tag. We found that treatment with HEP alone had a minimal impact on NHEJ-mediated guide-seq tag integration, with only a slight shift towards MMEJ repair events. In contrast, inhibition of DNA-PK resulted in near complete elimination of tag integration and a large shift toward MMEJ. Together, these results indicate that treatment with HEP has minimal impacts on NHEJ, off-target editing, and translocations making it ideal for use in research developing therapeutic applications where genome instability is of particular concern. Alternatively, for RUO applications, Alt-R HDR Enhancer V2 remains a potent facilitator of genome editing, with the combination of HEP and V2 offering the potential to further boost HDR rates.
Alt-R HDR Enhancer Protein does not change the off-target editing profile of Cas9
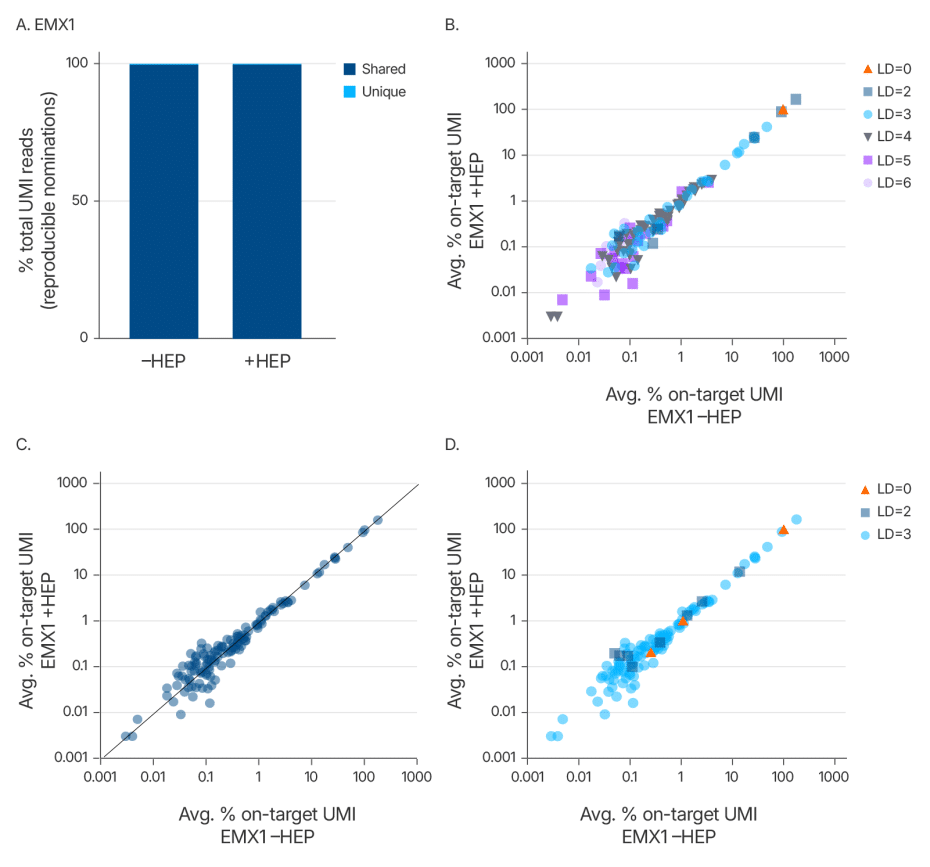
Maintaining editing specificity is critical for genome editing applications. To further evaluate whether the Alt-R HDR Enhancer Protein alters Cas9 off-target editing, we conducted an unbiased genome-wide off-target analysis using UNCOVER-seq in HEK293 cells stably expressing Cas9. Cells were treated with a guide RNA targeting the EMX1 locus and co-delivered with or without the enhancer protein. Off-target profiles were assessed based on unique molecular identifiers (UMIs) and categorized by Levenshtein distance, read correlation, and a tiered classification system that reflects predicted biological impact.
The analysis revealed no new off-target sites in the presence of the enhancer (Figure 5). Off-target frequencies and distributions were nearly identical between treated and untreated groups, confirming that the Alt-R HDR Enhancer Protein does not alter Cas9 off-target editing. These findings support the use of this enhancer in high-fidelity genome editing applications where preserving specificity is essential.
Alt-R HDR Enhancer Protein does not induce cellular toxicity across a wide concentration range
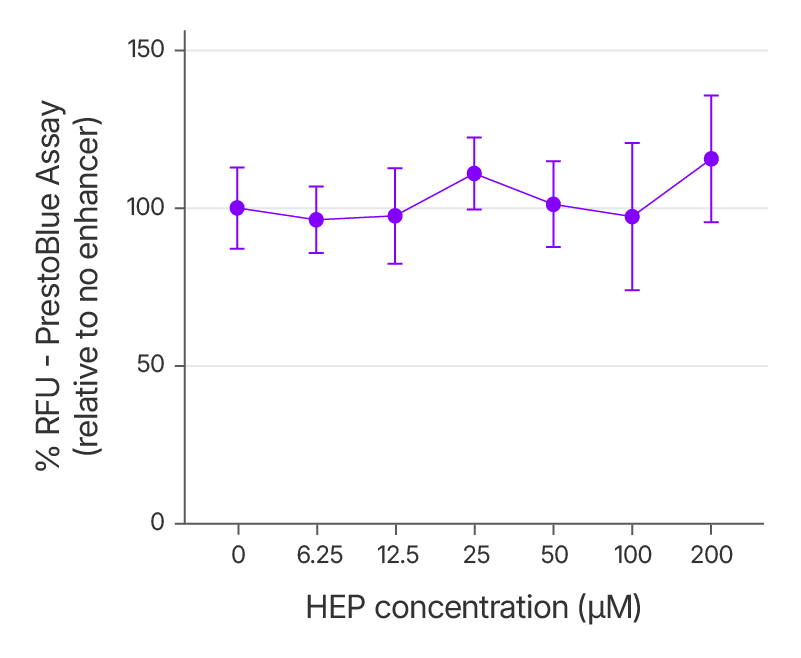
For any genome editing reagent intended for translational use, a favorable cytotoxicity profile is crucial. To assess this, we evaluated the effect of increasing concentrations of the Alt-R HDR Enhancer Protein on cell viability in HEK293 cells. Cells were treated with RNPs targeting EMX1, along with a single-stranded HDR donor and varying doses of the enhancer. Viability was measured 48 hours post-treatment using the PrestoBlue assay.
The results confirmed that even at elevated concentrations, the enhancer did not reduce cell viability relative to untreated controls (Figure 6). These findings demonstrate the protein’s excellent tolerability.
Together, these data establish the Alt-R HDR Enhancer Protein as a robust and reliable tool for improving the precision and efficiency of CRISPR-mediated genome editing. Across multiple genomic loci and cell types, including hard-to-edit cells such as human iPSCs, the enhancer consistently increased perfect HDR rates without compromising genomic integrity or cell viability. Importantly, the protein achieved these gains without increasing off-target indels, altering the off-target editing profile of Cas9, or elevating translocation frequency. Its favorable safety profiles make it particularly well-suited for high-fidelity editing workflows in both research and translational settings. By promoting HDR through a mechanism-specific pathway while preserving compatibility with existing delivery systems, the Alt-R HDR Enhancer Protein provides researchers with a powerful and flexible solution for next-generation genome engineering.
Explore additional data, protocol, and ordering options here.
References
- Canny MD, Moatti N, Wan LCK, et al. Inhibition of 53BP1 favors homology-dependent DNA repair and increases CRISPR-Cas9 genome-editing efficiency. Nat Biotechnol. 2018;36(1):95-102. doi:10.1038/nbt.4021
- Wang J, Sadeghi CA, Frock RL. DNA-PKcs suppresses illegitimate chromosome rearrangements. Nucleic Acids Res. 2024;52(9):5048-5066. doi:10.1093/nar/gkae1403.
- Cullot G, Aird EJ, Schlapansky MF, et al. Genome editing with the HDR-enhancing DNA-PKcs inhibitor AZD7648 causes large-scale genomic alterations. Nat Biotechnol. Published online November 27, 2024. doi:10.1038/s41587-024-02488-6
- Panier S, Boulton SJ. Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol. 2014;15(1):7-18. doi:10.1038/nrm3719
- Kinney KJ, Jia K, Zhang H, et al. UNCOVERseq Enables Sensitive and Controlled Gene Editing Off-Target Nomination Across CRISPR-Cas Modalities and Systems. Published online May 11, 2025. doi: https://doi.org/10.1101/2025.05.09.653165
For Research Use Only. Not for use in diagnostic procedures. Unless otherwise agreed to in writing, IDT does not intend these products to be used in clinical applications and does not warrant their fitness or suitability for any clinical diagnostic use. Purchaser is solely responsible for all decisions regarding the use of these products and any associated regulatory or legal obligations.
RUO25-3831_001