Alt-R™ CRISPR-Cas9 Libraries
Perfect for researchers looking for flexibility in their Cas9 guide design and custom plate configurations for CRISPR screening applications. IDT’s CRISPR gRNA libraries offer a highly customizable solution that can be integrated with analysis of editing sites, using the rhAmpSeq™ CRISPR Analysis System, for on- or off-target confirmation.
Custom libraries for better CRISPR screening.
Overview
Alt-R™ CRISPR-Cas9 Libraries
Flexible. Fast. Designed for Discovery.
Accelerate your gene editing research with Alt-R™ CRISPR-Cas9 Libraries—engineered for maximum flexibility, speed, and compatibility with your lab's automation workflows. Avoid the time-consuming steps of cloning and sequencing associated with lentiviral screens and get straight to results.
Why Choose Alt-R™ CRISPR-Cas9 Libraries?
- Intuitive Design Tool: Easily design CRISPR libraries for human and mouse genomes using our streamlined interface. Create your own gene list or select our predesigned targets
- Predesigned Libraries: Gene lists for human and mouse including druggable genome, kinases, proteases etc. See below for all available panels
- Project Dashboard: Save, review, and reorder your completed CRISPR library designs anytime.
- Integrated Analysis Tools: Your library designs integrate seamlessly into the rhAmpSeq design tool to evaluate on- and off-target editing.
- Flexible Library Options: Choose from a wide range of customizations: crRNA or sgRNA, plate types and formulations for your specific automation workflow
- Custom Plate Layouts: Upload your own plate configurations - supporting single or multiple guides per well.
- Expert Support: Get help with custom designs from our team of PhD-level scientists. Contact us.
Unlock the full potential of your gene editing projects—discover the top 5 reasons why CRISPR libraries from IDT are the smart choice.
Design your library
Easily enter or upload your human or mouse gene symbols into the design tool and within minutes we'll do the design work for you
Start designingUpload & order plates
Already have your sequences? Use the template to upload them and easily order in minutes
Upload sequencesRequest a consultation
Need design help or something custom? Our PhD experts are happy to help!
Get design helpProduct details
Alt-R™ CRISPR-Cas9 Libraries were developed to address the need for better CRISPR screening solutions. They are chemically modified guide RNAs synthesized on IDT’s proprietary, high-fidelity RNA manufacturing platform to provide high quality, reliable CRISPR-Cas9 libraries with fast delivery. If you don’t see what you need below you can contact us for custom options.
| Feature | Options |
|---|---|
| Design | Cas9 design available with design tool, custom and user-provided designs also accepted |
| Guaranteed yield | 0.1–10 nmol per oligo, delivered dry or formulated |
| Cas9 gRNA formats |
sgRNA
crRNA (without tracrRNA) |
| gRNA lengths supported | 19–20 nt (sgRNA or crRNA) |
| Chemical modifications | 2’-O-methyl RNA, PS linkages, end-blocking Alt-R modifications |
| Plate Types | PCR (96 well), V-Bottom (96 or 384 well), Deep Well (96 or 384 well) |
| Formulation types |
Arrayed format, multi-guide per well (pooled by gene) Arrayed format, single guide per well Custom formulations upon request |
| QC | Individual ESI/MS |
| Buffers |
RNase-free water
IDTE Buffer pH 7.5 |
| Quantity | 0.1–10 nmol per oligo |
| Volume | 20–800 µl |
| Concentration | 0.125–500 µM |
Not seeing what you need? Contact us for custom library options.
Alt-R CRISPR Enzymes
Our Alt-R CRISPR Enzymes are available in a variety of formats, with stock sizing available up to 50 mg. Larger formats and lot matching are also available for all products upon request.
| Cas protein | Available versions | Key features |
|---|---|---|
| S.p. Cas9 Nuclease V3 | Wild-type, 50% Glycerol Wild-type, Glycerol-free High Fidelity (HiFi) Wild-type Fluorescent-fusion | Targeting GC-rich regions
Low viscosity for high-throughput applications Reduced off-target activity* Fluorescent label for enrichment (GFP or RFP) |
| A.s. Cas12a (Cpf1) V3 | Wild-type Ultra |
Targeting AT-rich regions Increased on-target activity* |
| L.b. Cas12a (Cpf1) | Ultra | Increased on-target activity and low-temperature tolerance* |
* when compared to corresponding Wild-type controls
Predesigned libraries are available through the Alt-R CRISPR-Cas9 design tool, simply select one of the libraries below from the gene list drop down
| Human predesigned libraries | Gene Count |
|---|---|
| Druggable Genome | 5,657 |
| Drug Targets | 3,584 |
| Transcription Factors | 1,627 |
| Ubiquitin Enzymes | 848 |
| Ion Channels | 472 |
| Proteases | 475 |
| Protein Kinases | 545 |
| GPCRs | 387 |
| Phosphatases | 254 |
| Mouse predesigned libraries |
|---|
| Druggable Genome |
| Drug Targets |
| Transcription Factors |
| Ubiquitin Enzymes |
Turn around time*
| Product | Plate Type | # Total Oligos | Shipped (BD*) |
|---|---|---|---|
| Alt-R™ CRISPR-Cas9 crRNA Library Plate | 96 well | 96–384 | 7 |
| Alt-R™ CRISPR-Cas9 sgRNA Library Plate | 96 well | 96–384 | 9 |
| Alt-R™ CRISPR-Cas9 crRNA Library Plate | 384 well | 385–1536 | 9 |
| Alt-R™ CRISPR-Cas9 sgRNA Library Plate | 384 well | 385–1536 | 13 |
*BD = business days. In a few cases, wet plates (on dry ice) may automatically adjust to the next shippable day.
Product data
Alt-R™ CRISPR-Cas9 Libraries and CRISPR products offer an efficient solution for creating knockout libraries
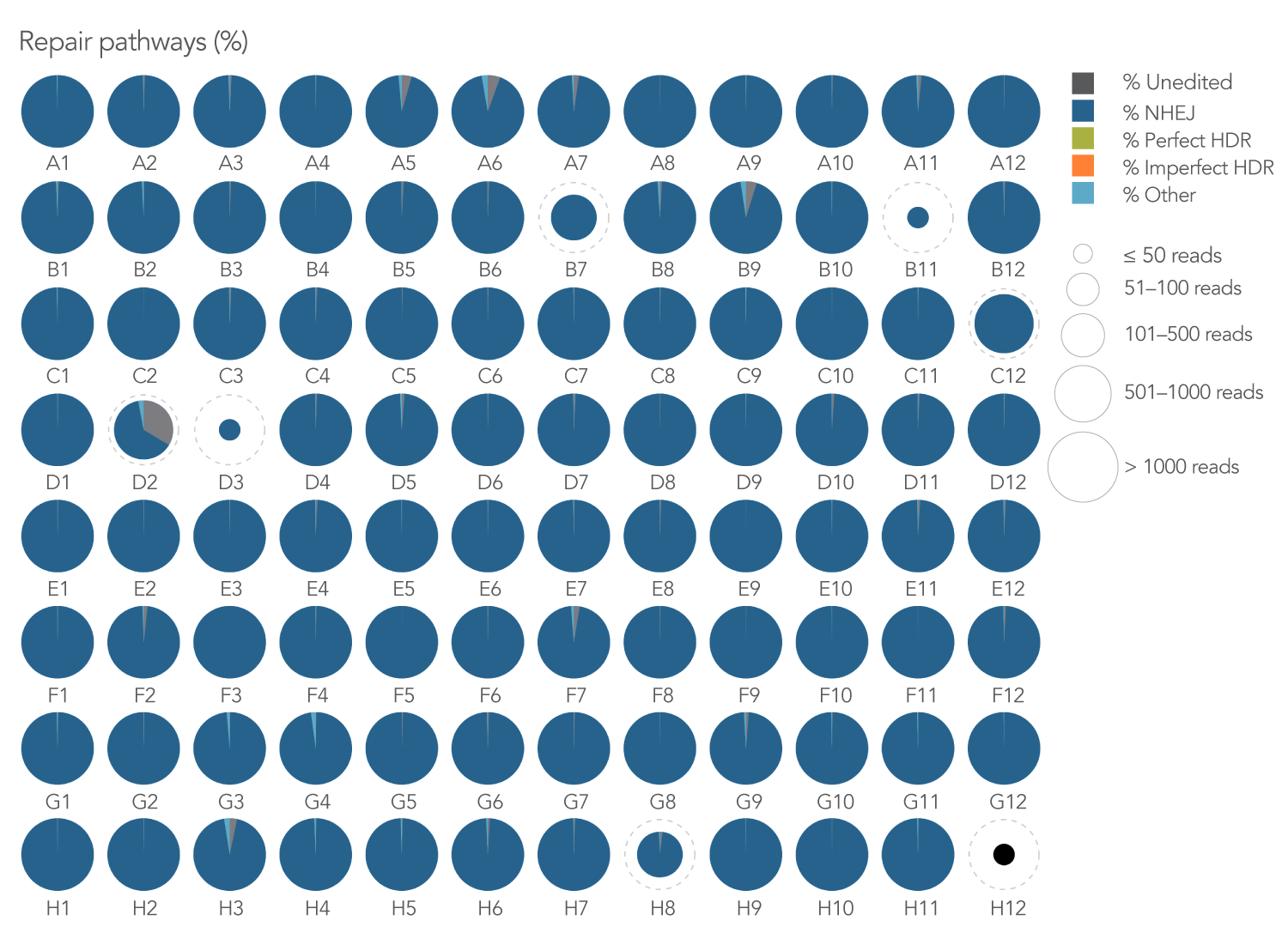
We investigated the efficiency of inducing non-homologous DNA end joining (NHEJ) by designing 3 sgRNAs per gene (N=95 genes) via an internal pipeline and using IDT’s Alt-R Cas9 nuclease S.p. Cas9 WT V3, electroporation enhancer, and rhAmpSeq CRISPR analysis system. The combined use of the Alt-R guides and additional CRISPR products provided an effective, high-throughput editing solution with >90% NHEJ editing in 99% of targeted genes (Figure 1).
Figure 1. A 3-guide-per-gene library design resulted in >90% NHEJ editing in almost all (90/91) targeted amplicons.
95 genes of interest were fed into an internal design pipeline to generate 3 guide RNA designs per gene, all
located within a 500 base span. Once target sites were identified, singleplex genotyping assays for each gene were selected using the rhAmpSeq Design Tool. K562 cells were transfected with 3 sgRNAs complexed to Alt-R S.p. Cas9 WT Nuclease V3 per well
(final concentration = 1 µM per guide). rhAmpSeq CRISPR Library preparation and subsequent sequencing on the Illumina MiSeq platform was then performed. IDT’s rhAmpSeq Analysis Tool revealed a high level of NHEJ-type editing events (dark
blue) in nearly all amplified regions (N = 91 amplicons with sequencing reads >500).
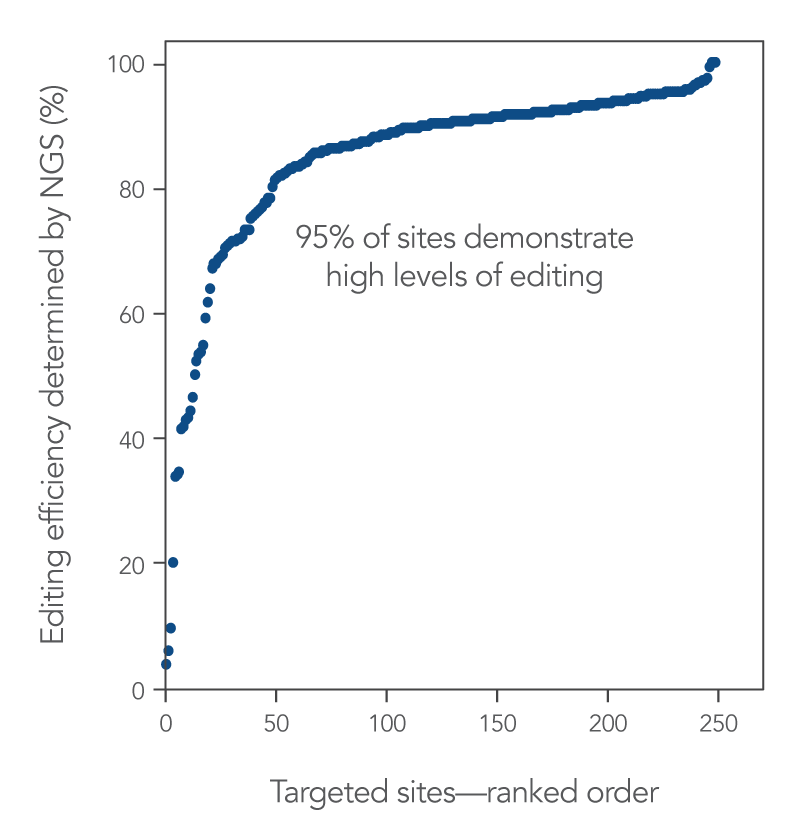
Alt-R CRISPR-Cas9 sgRNAs provide effective editing in Jurkat cells
To highlight the editing efficiency of sgRNAs, we designed sgRNAs targeting 255 sites across the human genome and delivered them to Jurkat cells (a human T-lymphocyte-derived cancer cell line) along with Alt-R S.p. WT Cas9 Nuclease V3. The result of this experiment shows that Alt-R CRISPR-Cas9 sgRNAs provide high levels of editing.
Figure 2. High levels of editing with Alt-R CRISPR-Cas9 sgRNAs.
Ribonucleoprotein (RNP) complexes were formed with Alt-R S.p. WT Cas9 Nuclease V3, combined with Alt-R Cas9 sgRNAs synthesized for 255 randomly selected Cas9 guide RNA sites across the human genome. RNP complexes (4 μM) were delivered into Jurkat cells via a Nucleofector™ system (Lonza) in the presence of Alt-R Electroporation Enhancer. Genome editing efficiencies were determined by target amplification followed by next generation sequencing (NGS) on an Illumina™ instrument.
Resources
Frequently asked questions
How do I reorder a previous gRNA library design?
A returning customer will have access to their CRISPR library design dashboard with unique DesignRun IDs for each of their completed designs. They can be reordered directly through your dashboard.
Can I order double stranded sequences so I can clone them into a vector?
No, Alt-R CRISPR-Cas9 libraries only allow single stranded crRNAs/sgRNAs. Double stranded sequences can be ordered as duplex DNA.
Can you accommodate different spacer lengths?
The spacer length must be between 19–20 nt for CRISPR libraries. Customers needing different spacer length could order them as custom gRNAs by filling out legacy "gRNA Plates iso 9001" or "gRNA Plates with Replicates ISO 9001", available through our custom order form configurator or discuss with one of our experts.
Can you accommodate DNA and RNA bases in a gRNA pooled format?
Only RNA bases are permitted on CRISPR library products. gRNA pools with DNA and RNA bases can be ordered as custom gRNA plate products.
Are additional customization options possible?
Yes, additional customization options are also available. Libraries with customized gRNA format, Cas9 alternative systems, yields, plate type, or formulation options that are not available through web ordering can be requested via custom request.
What are the different CRISPR Library formats available from IDT?
IDT offers gRNA libraries in multiple different formats. See product details for more information.
Do I need to perform an IVT reaction on my library?
The synthetic gRNAs are already an RNA molecule. Therefore, there is no need for in vitro transcription or cloning into an expression plasmid to generate a gRNA. The synthetic gRNA can be directly combined with Cas9 protein to prepare an RNP complex, ready to be delivered into cells for gene editing.
What type of CRISPR gRNA Libraries does IDT offer?
IDT offers synthetic gRNA libraries in many different formats, and they can range from tens to thousands of gRNAs in crRNAs or sgRNAs formats.
Do you have a whole genome predesigned library, i.e. Brunello?
Whole genome predesigned libraries are available for human and mouse genomes and can be requested by filling out the custom design intake form.
How were the targets for the predesigned libraries determined?
Gene pathway lists were derived from an IDT internal bioinformatic system. All other targets were designed against the entire protein-coding genome (when designs are possible given constraints) as defined by NCBI.
Can IDT provide sequencing services to assess for on- and off-target editing?
Yes, please visit our CRISPR analysis services page for more information on off-target nomination. CRISPR libraries are also integrated with the rhAmpSeq CRISPR analysis system for on- and off-target confirmation after editing.
Can I generate a library that targets introns?
No. Our CRISPR libraries design tool generates designs targeting only CDS (protein coding region). These can be requested as a custom library.
Can I design libraries for CRISPRa or CRISPRi? Can IDT do a custom design for this?
We do not currently support design of CRISPRa or CRISPRi libraries in our tool or by custom request.
Can the design output file from the design tool confirm exons targeted and/or exons included?
The design output file does not list targeted exons but does include target genes.
What exons are generally targeted by the tool?
The CRISPR Library Design tool preferentially targets early exons (within 60% of the longest transcript) that are most commonly represented across the canonical /annotated transcripts for the gene.
Do you offer NGS oligo design to analyze CRISPR editing?
Yes, we offer rhAmpSeq primer panel designs compatible with CRISPR library designs. Request for rhAmpSeq design can be submitted with our CRISPR library design tool.
What is the turnaround time for a custom library design?
Most of the designs are completed in 2 business days.
Does IDT offer custom CRISPR library design?
Yes, IDT offers design services if your designs are not able to be generated in our CRISPR Libraries design tool.
When should I use an HPLC-purified guide RNA instead of standard desalt?
Standard dealt purification is suitable for routine laboratory experiments, such as initial screening or proof-of-concept studies, where ultra-high purity is not as critical. We recommend you consider HPLC purification for applications where high specificity and efficiency are critical, such as in vivo experiments or translational applications.