Minimal residual disease (MRD) research
Tomorrow’s cancer research breakthroughs are on the horizon. The xGen MRD solution offers a complete sample preparation workflow including custom MRD hybridization capture panels delivered quickly and affordably.
xGen™ NGS—made for cancer research.
Overview
- More complete workflow—curated workflow for custom MRD hybridization research panels
- Rapid turnaround time—proven TAT of 5 business days from when order is placed
- Flexible panel size—target tumor-specific variants using up to 2000 probes per panel and order up to 50 panels at once
- Affordable MRD research—xGen MRD Hybridization Panels deliver a custom solution for under $40 USD per sample
- Rare variants—identify variants at ≤1% variant allele frequency (VAF) with your custom panel when paired with the xGen™ cfDNA and FFPE DNA Library Prep Kit
- Automation friendly—easy workflow that can be automated for high-throughput applications
- PCR analysis options—oPools™ Oligo Pools accommodate multiplexed PCR workflows for MRD research
Order now Request a consultation
Minimal residual disease (MRD) refers to the presence of cancer at low levels [1]. MRD research using NGS requires a full solution that encompasses initial variant discovery from individual tumors followed by custom target enrichment of those variants from low-abundance cell-free DNA (cfDNA) to identify the presence of circulating tumor DNA (ctDNA), indicative of MRD.
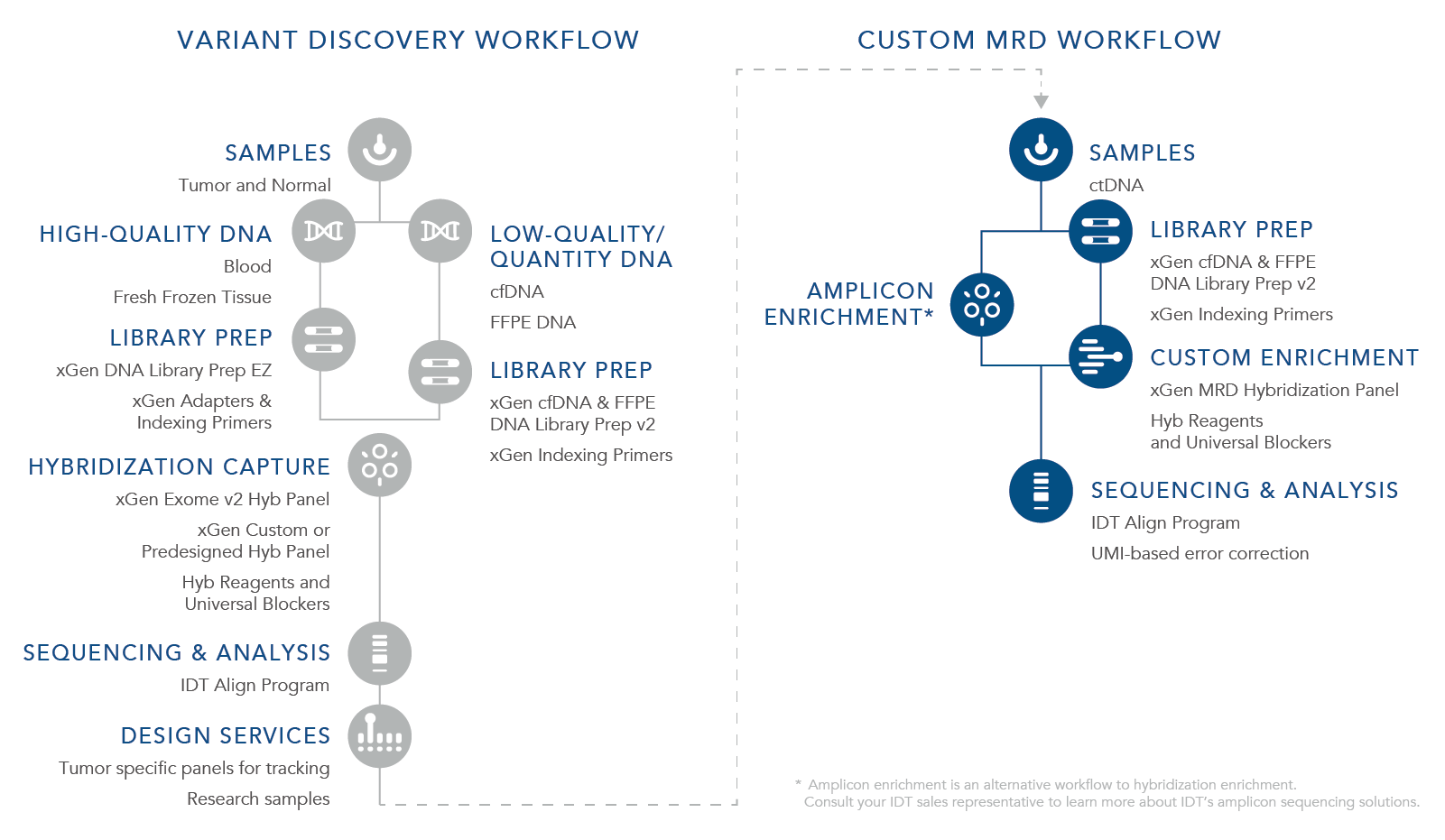
IDT offers a tumor-informed research workflow that first utilizes a custom or predesigned hyb capture panel with a broad target region to identify variants that are representative of each specific cancer (Table 1). Our panels are designed using a proprietary artificial intelligence (AI) algorithm that reduces off-target binding while maximizing coverage for identifying variants. Learn more about the design algorithm in this white paper The xGen™ Off-Target Quality Control (QC) Method increases accessible probe design space for hybridization capture when compared to the repeat annotation based QC method.
Table 1. Variant discovery workflow.
| Step | Benefits | ||
|---|---|---|---|
| Samples | Tumor and Normal | ||
| Sample Type | Blood or Fresh Frozen Tissue | cfDNA or FFPE DNA | |
| Library prep | xGen DNA Library Prep Kit EZ or xGen cfDNA & FFPE DNA Library Prep Kit v2 | xGen cfDNA & FFPE DNA Library Prep Kit v2 | Flexibility in choice of library preparation kits to accommodate a range of sample types |
| Target enrichment | Create an xGen Custom Hyb Panel or use one of the IDT Predesigned Hyb Panels, such as the xGen Exome v2 Hyb Panel or the xGen PanCancer Hyb Panel | AI- generated hybridization probe design maximizes coverage of targeted regions | |
| Sequencing & Analysis | IDT Align Program | Minimizes your search time to find a sequencing provider tailored to your research | |
The identified variants become the targets for a customized tumor-specific xGen MRD Hybridization Panel that can be designed, synthesized, and shipped within 5 business days from when the order is placed. The recurrent use of this panel with deep sequencing can support researchers in monitoring the potential presence or absence of targeted biomarkers at ultra-low frequency (Table 2).
Since each panel is used to identify rare alleles within a cfDNA sample, a high sample conversion rate is essential during library prep. With the xGen cfDNA & FFPE DNA Library Preparation Kit, low-frequency tumor-specific mutations can be investigated when used in conjunction with customized MRD Panels. Enrichment of the specific variants using a custom xGen MRD Hyb Panel furthers the identification of ultra-low variants by focusing the library on a small target space for the required deep sequencing.
Table 2. Custom MRD research workflow.
| Step | Benefits | |
|---|---|---|
| Design | Tumor-specific panel design | Design up to 50 unique panels in one order |
| Library prep | xGen cfDNA & FFPE DNA Library Prep Kit v2 | Higher conversion rate than competitor offerings |
| Target enrichment | xGen MRD Hyb Panel(s) | Enrichment of tumor-specific mutations for deep sequencing and identification of ultra-low variants |
| Sequence & Analysis | IDT Align Program | Minimizes your search time to find a sequencing provider tailored to your research |
As an alternative, IDT also offers oPools™ Oligo Pools that support multiplex PCR workflows for MRD research. oPools Oligo Pools include up to 20,000 pooled oligonucleotides that can be from 40–350 bases in length. These panels can be used to generate amplicon sequencing libraries for MRD research (see the workflow graphic).
Method data
Variant discovery workflow followed by the xGen Custom MRD Workflow
Researchers use biomarkers to track ctDNA found in circulating cfDNA, and/or formalin fixed paraffin embedded (FFPE) samples to research cancer. Initial analysis involves identifying the specific somatic variants found in a sample. Common MRD research approaches for identifying such variants involve preparing libraries from extracted cfDNA and FFPE, then performing hybridization capture using larger custom or predesigned panels with a broad number of targets to find unique signatures for each research sample. This approach is used during the early research stage and discovery phase to identify somatic and germline mutations, but due to the larger size of the panel and associated sequencing depth required, it is not used to identify ultra-low variants in MRD research samples.
For rare variant discovery, IDT offers an xGen Custom MRD Workflow to assist you in your MRD research needs. Easily prepare high complexity libraries from extracted cfDNA using the xGen cfDNA & FFPE DNA Library Prep Kit. For target enrichment and accurate ultra-low variant identification, design your xGen MRD Hybridization Panels with up to 2000 probes per panel and order up to 50 panels at once. These customizable panels can enable reliable variant identification at ≤1% variant allele frequency (VAF) if input cfDNA quantity and sequencing depth requirements are met (see Figure 3).
Comprehensive conversion and error correction enables ultra-low variant identification with cell-free DNA
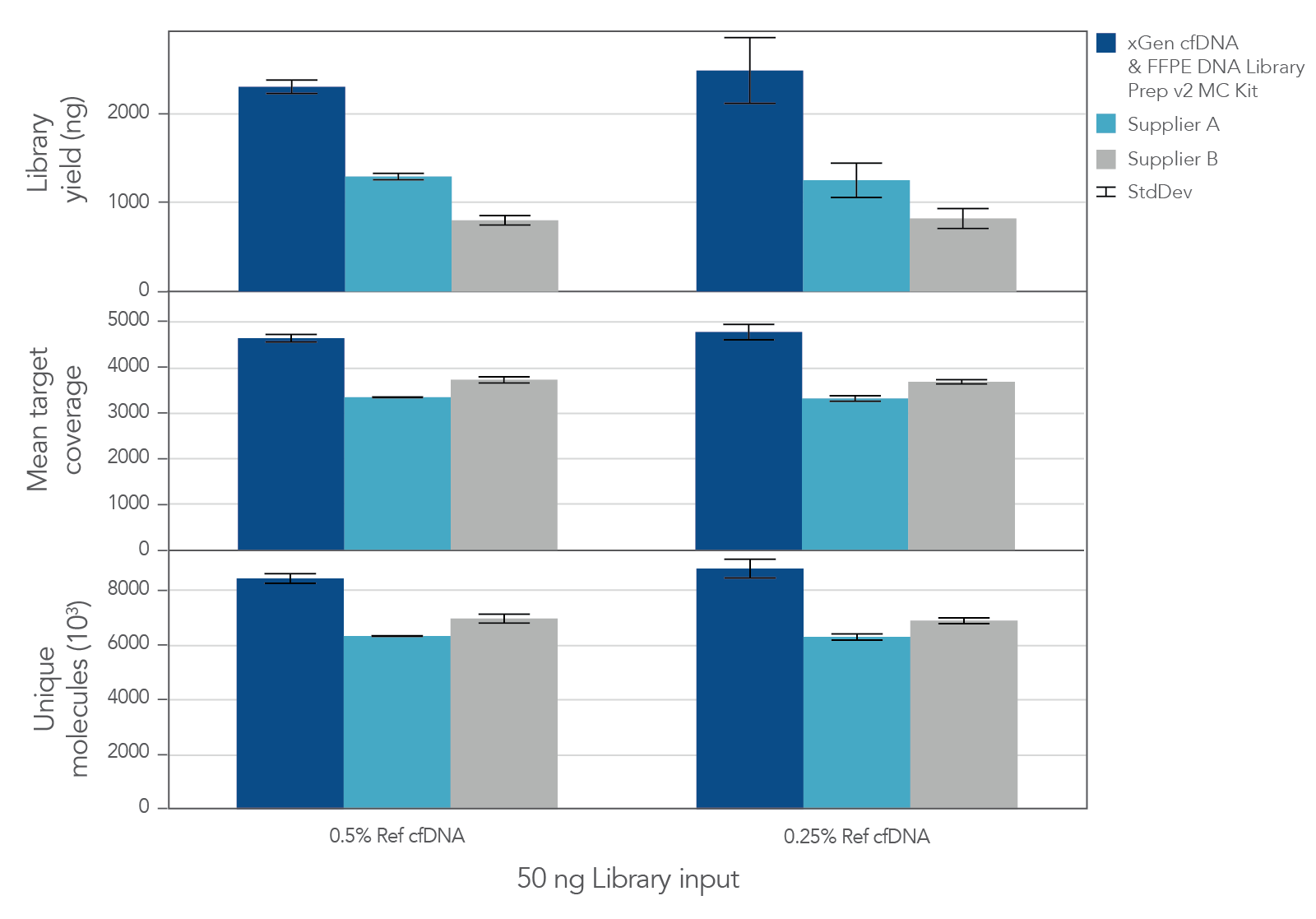
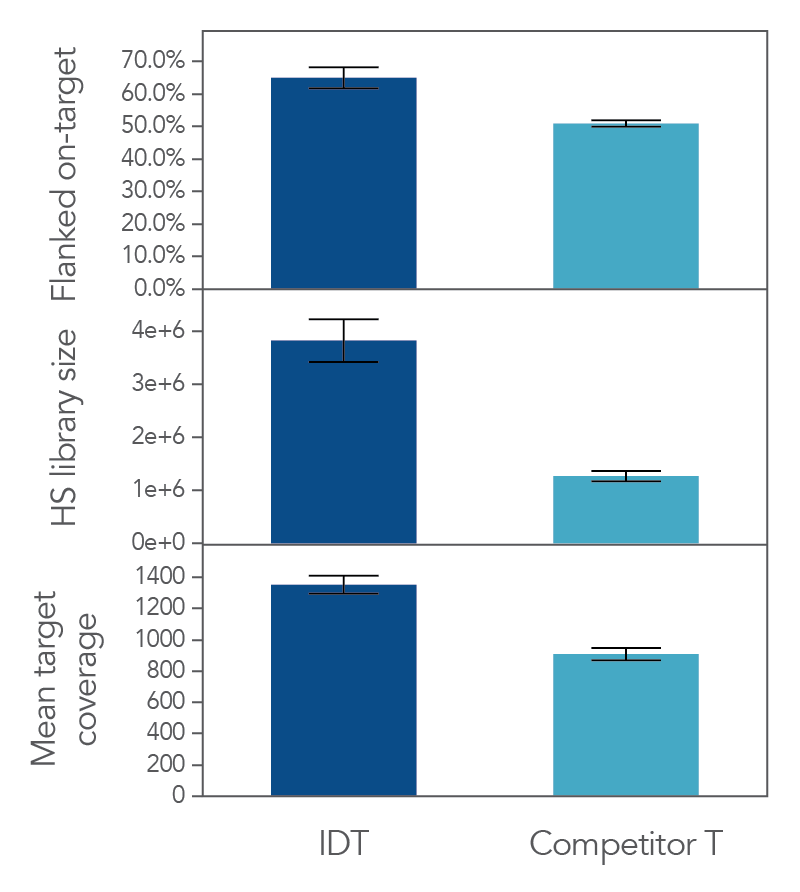
The unique, single-stranded ligation strategy of the IDT xGen cfDNA & FFPE DNA Library Prep Kit paired with the xGen hybridization capture workflow delivers high conversion of input DNA molecules to sequencing data. This high conversion rate is critical for identification of ultra-low frequency variants, which is common in the analysis of cfDNA samples. Whether comparing the xGen cfDNA & FFPE DNA Library Prep Kit against competing library prep kits within the xGen Hyb Capture Workflow (Figure 1) or the complete IDT and competitor T workflows against each other (Figure 2), we observe a noticeable difference in library complexity and mean target coverage in xGen Hyb Capture Workflow. This advantage translates into increased cost savings given the lower sequencing requirements needed to hit certain minimum coverage depth thresholds.
Figure 1. xGen cfDNA & FFPE DNA Library Prep MC Kit delivers higher yields, complexity, and coverage. Libraries were prepared in triplicate according to the manufacturer’s instructions with 50 ng of Horizon cfDNA reference standard and 7 cycles of PCR. Following quantification, libraries were captured with a 61 kb (target space) xGen Custom Hyb Panel using the xGen Hybridization and Wash Kit. Captured libraries were pooled and sequenced on a NextSeq 500 (Illumina) using a high output 300 cycle kit and the manufacturer's protocol. After subsampling to 85M total reads, coverage and complexity were calculated.
Figure 2. xGen cfDNA & FFPE library prep produces higher on target percentages, more complex libraries and deeper coverage that another commercial MRD solution provider in a hyb capture workflow. A third party was used to generate xGen cfDNA & FFPE or competitor T libraries using Covaris-sheared gDNA (Coriell), followed by custom hybridization capture using an Oncology panel designed by the respective vendors with a target space of 112kb. Sequencing was performed on an Illumina NovaSeq and subsampled to 7 million reads/ sample (n = 24 per vendor). Data was analyzed using Picard and hg38.
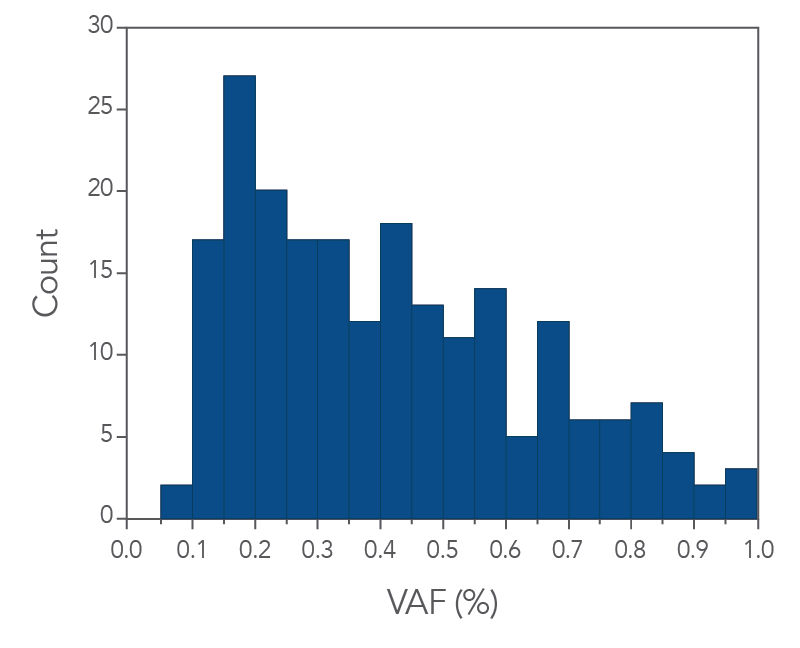
In addition, the xGen cfDNA & FFPE DNA Library Prep Kit includes adapters that contain unique molecular identifiers (UMIs), which enable bioinformatic error correction. Combining higher complexity and coverage with stringent error correction better enables the identification of ultra-low frequency variants (Figure 3 below). As shown in Figure 3, the xGen MRD workflow confidently identifies variants below 1% and can even enable identification of ultra-low variants at or below 0.1% VAF using as little as 10ng of cfDNA input for library preparation.
Figure 3. IDT’s custom MRD Panels are able to capture low levels of VAF. The histogram shows the variant count for a given VAF bin. xGen cfDNA & FFPE DNA Libraries (n = 1) were made using 10 ng of cfDNA. Single-plex captures were performed with a custom MRD hyb panel (81kb) and a 16-hour hybridization. Sequencing was performed by using 2 x 150 PE reads on an Illumina NextSeq 550/500 platform and data was subsampled to 25 M total reads.
Ordering
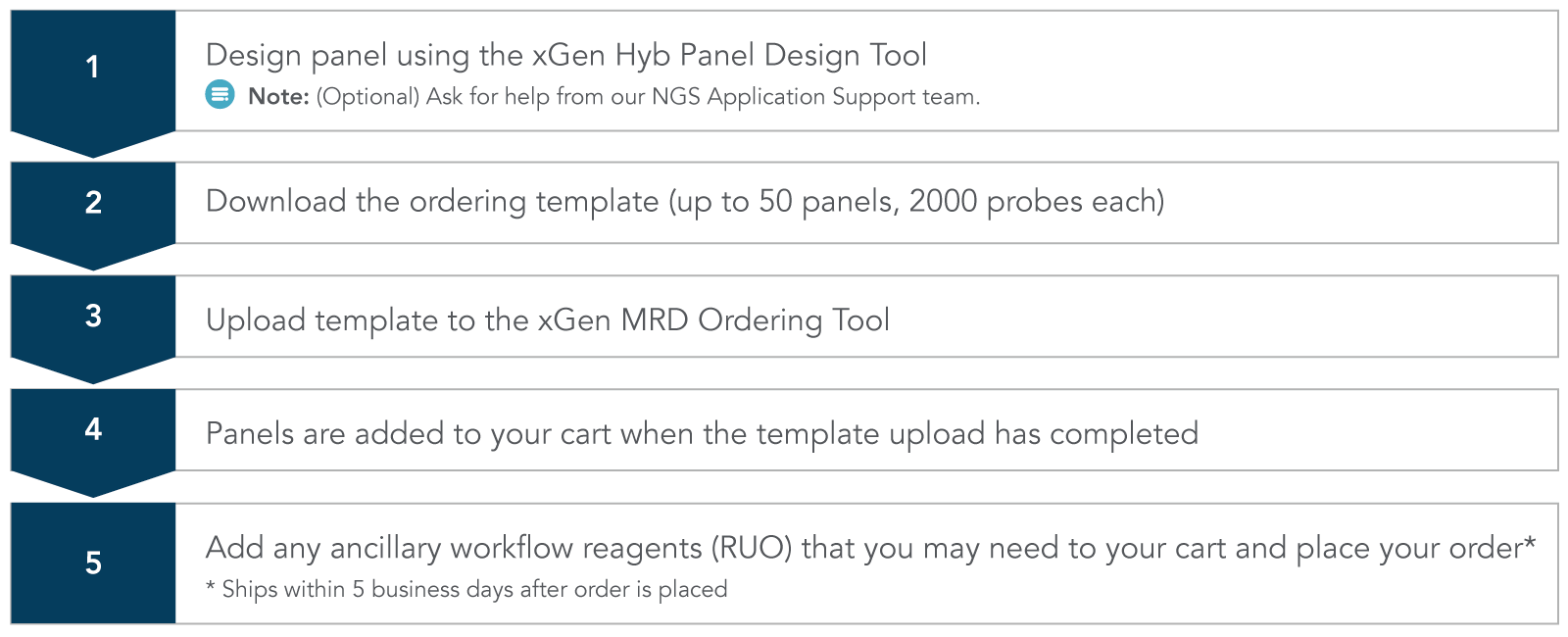
Simple 5 step ordering process
Custom MRD workflow research products
xGen MRD Hybridization Panel is optimized for use with the xGen Hybridization and Wash Kit, xGen Universal Blockers, and xGen Library Amplification Primer Mix, which can be ordered as ancillary RUO reagents part of the “Order my design” process below.
Create custom designs for xGen MRD Hyb Panels
Please contact the IDT NGS Applications Support team for assistance with processing your xGen MRD Hyb Panel Design.
Request supportOrder your predesigned panels
If you have predesigned probe sequences, upload them using our submission form.
Order my designRequest MRD workflow consultation
For more information on the MRD workflow solutions, please contact IDT sales.
Request sales consultationxGen Minimal Residual Disease (MRD) Hybridization Panel
Resources
References
- Thierry AR, El Messaoudi S, Gahan PB, et al. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 2016;35(3):347-376.