Minimal residual disease research
Overview
Minimal residual disease (MRD) refers to the presence of cancer at low levels. MRD researchers can use NGS to identify low levels of cancer that remains in circulating cell‑free DNA (cfDNA) by looking for the presence of low frequency somatic mutations already associated with the underlying tumor or cancer type.
What is minimal residual disease (MRD)?
When a low level of cancer remains, this is called minimal residual disease (MRD) and is a predictor of cancer relapse. Typically, prior to MRD monitoring, tumors are assessed for specific somatic or germline mutations underlying the disease, which then serve as highly specific markers to identify cancer during longitudinal research studies. Revealing MRD in circulating cell-free DNA (cfDNA) has been shown to predict relapse, however, one of the biggest challenges is that many samples have a very low amount of circulating tumor DNA (ctDNA) within the already low concentration of cfDNA. The mutation frequencies may be at 1% or lower and very difficult to detect. To obtain the best results, a research workflow is needed that will identify low frequency somatic alleles with confidence. By using next generation sequencing (NGS), minute fractions of ctDNA indicative of cancer can be identified to determine whether cancer has been completely eradicated [1]. Ideally, MRD identification would be both broadly applicable and highly selective, thus allowing for improved outcomes.
Minimal residual disease in cancer
MRD is very commonly used with blood cancers, such as leukemia, lymphoma, or multiple myeloma, as well as for other cancers such as lung cancer, colorectal cancer, and breast cancer. In cases of acute myeloid leukemia (AML), a positive MRD test generally results in a higher rate of disease recurrence and shorter survival rate [2]. Monitoring MRD levels for non-Hodgkin lymphoma (NHL) allows for more accurate assessments by pinpointing the degree of risk or allowing for earlier prevention of potential relapse [3]. By measuring the presence of ctDNA for lung and colorectal cancer, identification of tumor-specific mutations can occur months earlier than radiographic methods, when disease burden is lower and results are potentially more effective [4]. Metastatic breast cancer recurrence more than 5 years after the initial onset often results from micrometastatic disease that was there from the beginning but was not at identifiable levels via traditional imaging and blood testing. Revealing MRD in cfDNA has been shown to predict relapse with high accuracy allowing for early, more tailored responses making research in this area extremely important [5-6].
Importance of MRD research
All too often a small number of cancer cells remain and goes undetected until there are noticeable symptoms. By this time, it may be too late to effectively get rid of the disease and it may have already spread, leading to irreparable damage. The ability to track ultra-low frequency tumor-specific cancer mutations from cell‑free DNA as early as possible is an important advancement for cancer research. Research using the development of library preparation methods that maximize conversion of low abundance cfDNA samples combined with tumor-specific targeted deep sequencing methods aims to understand the lowest levels of cancer possible while still maintaining accuracy. Some of the current methods in the literature allow for detection as low as 0.001% tumor plasma cells using NGS sequencing [7]. NGS panels are also being developed to increase the types of cancers that can be identified using MRD, with the option of customizing the panel to account for the great variability in mutations that can be seen even within the same type of cancer. Research and development continues to improve the testing methods to push the levels of recognition even lower and expand the biomarkers that can be characterized to find ways to enhance the identification of MRD in samples.
ctDNA is a biomarker for the assessment of both MRD and the potential for recurrence. For a research workflow to be beneficial for MRD recognition and to be used with cfDNA, it is imperative that it can identify the lowest levels of residual cancer present. Currently there are two approaches for evaluating MRD using cfDNA from liquid tumor samples including tumor-informed and the custom MRD research workflow. A tumor-informed approach is informed by genomic sequencing of the primary tumor while the custom MRD research workflow is uninformed by the mutations in the primary tumor. With the custom MRD research workflow, a preselected panel of assays for select tumor mutations is used to sequence samples using a fixed set of genes looking for certain cancers or looking for several variants. This strategy is used during the research stages as well as the discovery phase, is not patient specific, and utilizes a stocked panel such as xGen™ Acute Myeloid Leukemia (AML) Cancer Hybridization Panel offered by IDT. The tumor-informed approach shifts to specific panels in which sequencing depth is important to find an exact mutation or mutational signature of a tumor that can then be used to design a custom sequencing panel specific to the tumor DNA being further researched. For the application, xGen MRD Hyb Panel and the xGen cfDNA & FFPE DNA Library Preparation Kit can provide the most benefit.
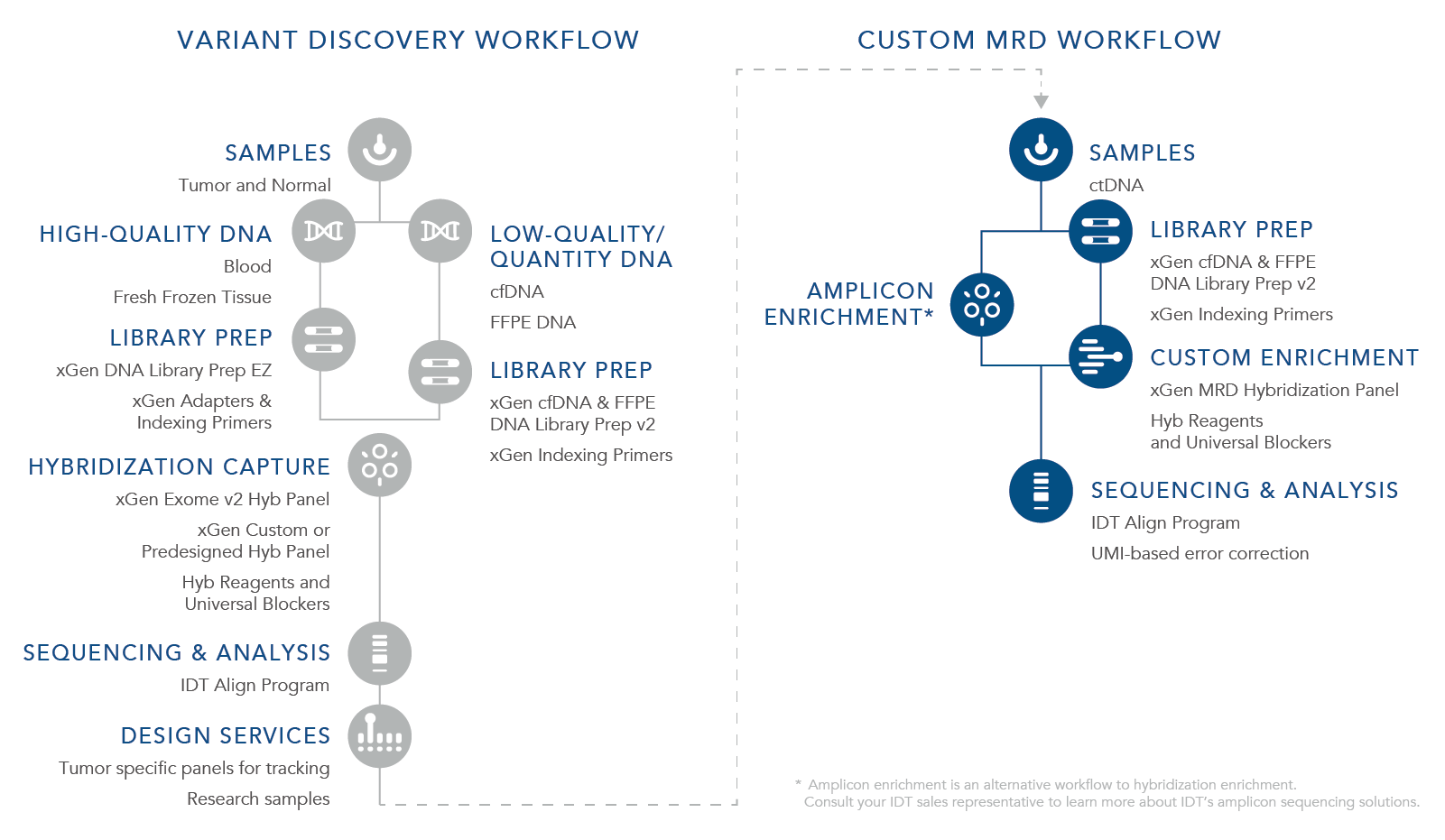
How do you measure MRD?
With an MRD positive test, there are still cancer cells present whereas with an MRD negative result, there are no cancer cells present, which has been correlated with longer remission periods and improved survival rates [8]. Levels of MRD can be measured in several different ways and often depends on the type of cancer. For AML flow cytometry, polymerase chain reaction (PCR), and NGS methods are used to determine MRD levels while for NHL researchers often use PCR and NGS [2-3]. One important issue with assessing MRD with multiple myeloma is that samples are taken from bone marrow, which may not provide a true representation of the actual tumor burden. Given the inaccurate results from this type of sample collection, MRD testing in this type of cancer is moving to magnetic resonance imaging as it is a more sensitive technique that provides an accurate report of MRD levels. This method cannot be used alone, however, as it cannot determine state of a lesion following therapy. Studies suggest this approach would need to be used in combination with other techniques [9]. Liquid biopsy detection of circulating tumor DNA (ctDNA) in blood plasma is often used for colorectal, lung, and breast cancer to determine MRD levels [4-6].
Challanges with MRD Identification
One of the biggest challenges with MRD Identification is that often samples have a very small number of cells that are very difficult to detect. To obtain the best results, is important for research efforts to find a test that will identify small populations of cells. In addition, there may be a mixed population of cells present, so the method selected needs to be deemed sensitive enough to identify these cell types. Often samples are collected from a liquid biopsy to study for MRD in a peripheral blood sample from cfDNA. Analyzing these samples can be difficult given they contain only a few thousand copies of each gene [5]. Use of the IDT xGen cfDNA & FFPE DNA Library Preparation Kit enables variant identification from degraded and low-input research samples which can allow for identification of even ultra-low frequency variants. Each cancer variant contains its own unique set of mutations making identification of cells more complicated. Overcoming this challenge requires the development of MRD assays with a heightened accuracy for low frequency variant detection to avoid false negative results and to ensure the identification of MRD before metastatic disease develops [5]. The xGen MRD Hyb Panel is an affordable solution with a fast turnaround time (after design acceptance) that can be customized to the target of interest.
Cancer molecular profiling
Research in the discovery and identification of new, targetable biomarkers is driven by comprehensive tumor profiling using NGS. However, converting tissue samples into NGS libraries is often challenging due to the low quantity and quality of DNA in such samples. Download this application note to explore how low-frequency variants have been identified in this application.
Applicable MRD research technologies
Products for MRD research
Explore IDT’s resources for MRD research
References
- Dogliotti I, Drandi D, Genuardi E, Ferrero S. New Molecular Technologies for Minimal Residual Disease Evaluation in B-Cell Lymphoid Malignancies. J Clin Med. 2018;7(9):288.
- Buccisano F, Hourigan CS, Walter RB. The Prognostic Significance of Measurable ("Minimal") Residual Disease in Acute Myeloid Leukemia. Curr Hematol Malig Rep. 2017;12(6):547-556.
- Chase ML, Armand P. Minimal residual disease in non-Hodgkin lymphoma - current applications and future directions. Br J Haematol. 2018;180(2):177-188.
- Chaudhuri AA, Chabon JJ, Lovejoy AF, et al. Early Detection of Molecular Residual Disease in Localized Lung Cancer by Circulating Tumor DNA Profiling. Cancer Discov. 2017;7(12):1394-1403.
- Parsons HA, Rhoades J, Reed SC, et al. Sensitive Detection of Minimal Residual Disease in Patients Treated for Early-Stage Breast Cancer. Clin Cancer Res. 2020;26(11):2556-2564.
- Garcia-Murillas I, Schiavon G, Weigelt B, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med. 2015;7(302):302ra133.
- Yao Q, Bai Y, Orfao A, Chim CS. Standardized Minimal Residual Disease Detection by Next-Generation Sequencing in Multiple Myeloma. Front Oncol. 2019;9:449.
- Short NJ, Zhou S, Fu C, et al. Association of Measurable Residual Disease With Survival Outcomes in Patients With Acute Myeloid Leukemia: A Systematic Review and Meta-analysis. JAMA Oncol. 2020;6(12):1890-1899.
- Hillengass J, Merz M, Delorme S. Minimal residual disease in multiple myeloma: use of magnetic resonance imaging. Semin Hematol. 2018;55(1):19-21.


