Affinity Plus™ qPCR Probes
Increase SNP target specificity and use shorter sequences with these high Tm locked nucleic acid probes
Use Affinity Plus™ qPCR Probes for enhanced discrimination of thermodynamically similar samples such as single nucleotide polymorphisms and transcript variants. The Affinity Plus bases used in these qPCR probes include up to 6 locked nucleic acid monomers (see the technology page, Locked nucleic acids, for more information). When incorporated into a probe, locked nucleic acids impart heightened structural stability, leading to increased hybridization melt temperature (Tm).
Ordering
- Select from a wide range of fluorophores including FAM, Yakima Yellow® (YAK), SUN™, HEX, Cy® 3, Cy® 5, TEX, and TYE™ dyes
- Improve mismatch discrimination compared to traditional qPCR probes
- Benefit from this affordable approach to increasing assay hybridization specificity
- Make flexible Tm adjustments
- Choose Mini Affinity Plus qPCR Probes for screen small sample sets
Prices listed include probe sequence (10−25 standard bases with up to 6 Affinity Plus LNA bases), reporter, quencher, and HPLC purification. See the Product Details section for design tips.
Mini Affinity Plus qPCR Probes
Mini Affinity Plus qPCR Probes are ideal for screening small sample sets, using on digital PCR platforms, or performing just a few reactions when optimizing probe designs. Guaranteed normalized yield of 0.5 nmol.
|
Affinity Plus qPCR Probes
Affinity Plus qPCR Probes are offered with a wider selection of dyes and quenchers than Affinity Plus Mini qPCR Probes, and are best-suited for large scale or high-throughput applications.
Cy is a registered trademark of Cytiva; Black Hole Quencher (BHQ) is a registered trademark of Biosearch Technologies, Inc.; TEX™ fluorophore is a trademark of ThermoFisher; TYE fluorophores are licensed from ThermoFisher; Yackima Yellow (YAK) is a registered trademark of EliTech Group
Product details
Advantages of Affinity Plus qPCR Probes containing locked nucleic acid modifications
Locked nucleic acid bases, such as Affinity Plus monomers, can be incorporated into effective qPCR probes [1-4]. Because locked nucleic acid bases significantly increase Tm, Affinity Plus qPCR Probes can be designed with shorter lengths than standard qPCR probes. Shorter probes have better quenching and a higher signal-to-noise ratio than unmodified longer probes. More importantly, locked nucleic acid probes offer an improved ability to distinguish mutations or single nucleotide polymorphisms (SNPs) [1]. An Affinity Plus qPCR Probe can be designed with several locked nucleic acid monomers resulting in a Tm of >15°C, which greatly increases accuracy of allele determination in real-time PCR or other methods that use differential hybridization to distinguish polymorphisms.
Design considerations
- Depending on sequence context, each locked nucleic acid base insertion into a DNA oligo will increase the Tm by 3−6°C. Multiple locked nucleic acid additions can result in an Affinity Plus qPCR Probe with a Tm of >15°C.
- The relative binding affinity (Tm) of Affinity Plus locked nucleic acid bases are Affinity Plus:Affinity Plus > Affinity Plus:DNA > DNA:DNA. Therefore, it is important to examine the probe sequence for self-dimer and hairpin formation and minimize designs that allow Affinity Plus:Affinity Plus pairing. Use the OligoAnalyzer™ Tool to check secondary structure predictions.
- Add up to 6 Affinity Plus locked nucleic acid bases in an Affinity qPCR Probe
- Affinity Plus locked nucleic acid bases should be placed at the SNP site and on each of the adjacent bases. The SNP should be positioned in the center of the probe, if possible. Avoid placing Affinity Plus locked nucleic acid bases on the first or last bases of the probe sequence.
- Additional Affinity Plus locked nucleic acid bases can be incorporated to adjust Tm as needed. We recommend placing no more than 4 Affinity Plus locked nucleic acid bases sequentially.
- Placement of the Affinity Plus locked nucleic acid bases is sequence dependent and may require optimization to reach the ideal Tm between match and mismatch.
- Amplicon sizes up to 150 bp are recommended for standard qPCR. Shorter amplicons of 80−100 bp are typically recommended for digital PCR (dPCR) or for applications with samples containing shortened fragments (such as FFPE or small circulating DNA samples).
Product data
Enhanced sequence stability
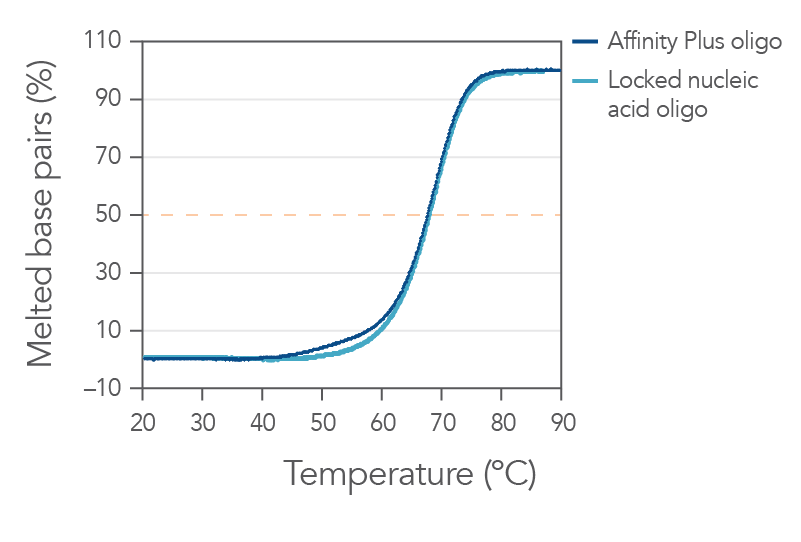
Sequences containing Affinity Plus locked nucleic acid nucleotides provide identical annealing properties as one other manufacturers' locked nucleic acid sequences (Figure 1). The incorporation of locked nucleic acid bases increases hybridization affinity, the melting temperature (Tm), and enhances sequence stability both in vitro and in vivo.
Figure 1. Sequences containing Affinity Plus show identical annealing properties to locked nucleic acid sequences from another manufacturer. The percentages of duplex melted base pairs are plotted as a function of temperature. The 15-mer sequence, GGTCCT+T+A+CTTGGTG, was synthesized with either Affinity Plus or other locked nucleic acid modifications incorporated at the +T, +A, and +C sites. These oligos were mixed with the complementary DNA strand (1 µM each strand) in 1 M Na+ buffer (pH 7). Melting curve analyses were performed as described in Owczarzy, et al [2]. The plots of duplex melted base pairs were averaged from at least 7 heating and cooling melting curves. Melting temperature was measured to be 67.7 ± 0.3°C for the Affinity Plus oligo and 67.9 ± 0.3°C for the other locked nucleic acid oligo. Free energy of duplex hybridization at 37°C was determined to be -18.1 ± 0.9 kcal/mol for the Affinity Plus oligo and -18.7 ± 0.9 kcal/moI for the other locked nucleic acid oligo. These results demonstrate the identical annealing properties of both sources of locked nucleic acid sequences.
Locked nucleic acid probe advantage in qPCR assays
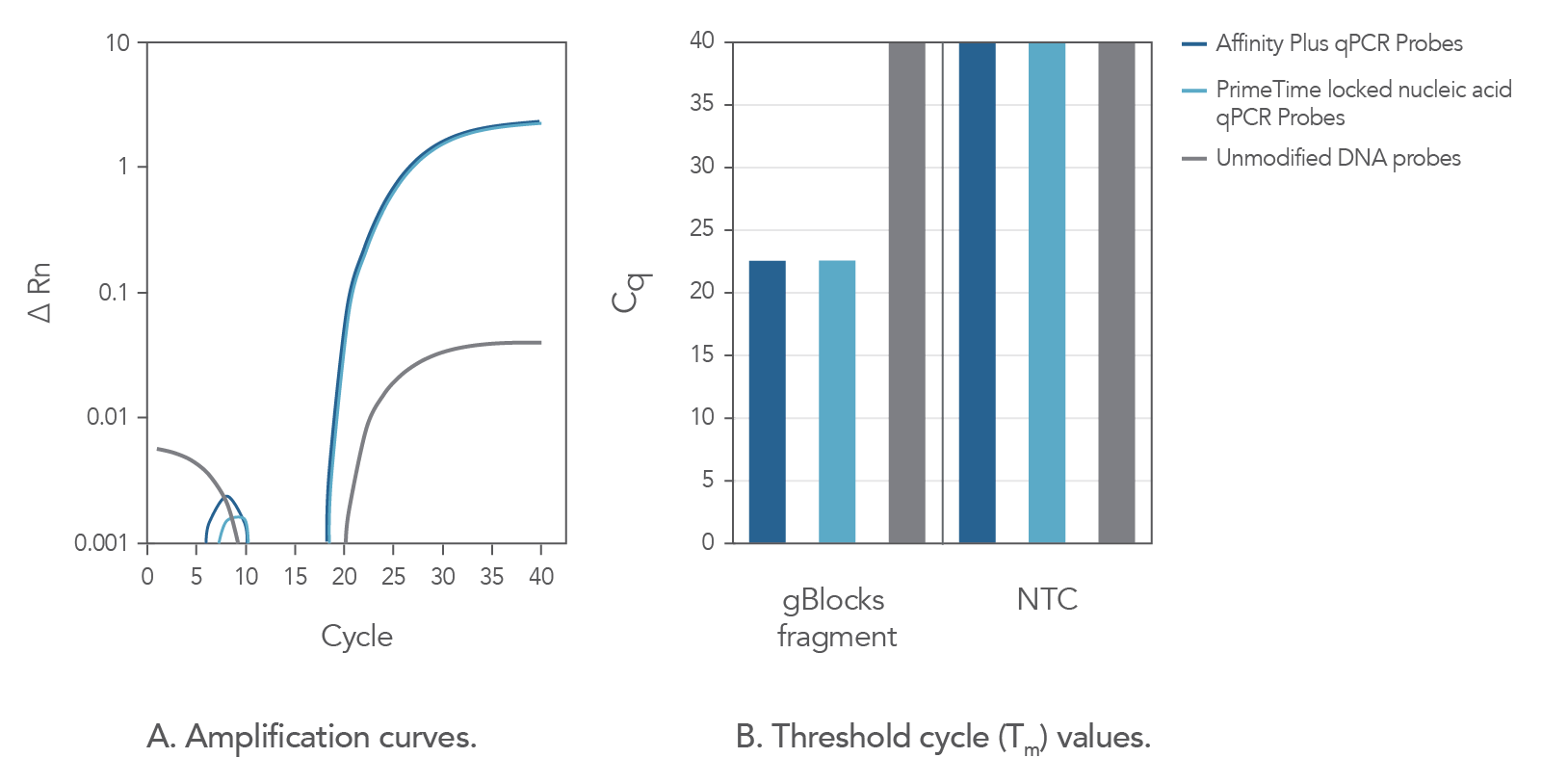
In comparison to unmodified probe sequences, Affinity Plus qPCR Probes have higher melting temperatures and are more stable in qPCR assays when targeting regions of low GC content. The increased stability also makes possible the use of shorter probe designs, ideal when target regions are limited in size. Figure 2 demonstrates that Affinity Plus qPCR Probes provide the same amplification profile as PrimeTime™ qPCR locked nucleic acid probes, with both showing more rapid target amplification vs unmodified DNA probes.
Figure 2. Affinity Plus qPCR Probes generate identical amplification data as PrimeTime Locked nucleic acid qPCR Probes. A custom PrimeTime qPCR assay was designed to detect the M2-2 gene from the Human respiratory syncytial virus A strain. Using a gBlocks Gene Fragment as a target template, the performance of 2 locked nucleic acid qPCR probes [17-base probes containing 7 locked nucleic acid monomers at identical positions, manufactured as either Affinity Plus qPCR Probes or PrimeTime locked nucleic acid qPCR Probes (Tm = 68.1°C)] were compared to the same DNA sequence without lock nucleic acid modifications (Tm = 49.9°C). Template and probe sequences are shown below in Table 1, (see Figure 2 sequences), with locked nucleic acid positions denoted as +N. (A) Amplification curves using the 3 probes reveal that the Affinity Plus and PrimeTime locked nucleic acid qPCR Probes yield identical results. (B) Threshold cycle (Cq) values comparing reactions with (gBlocksTM Gene Fragment) or without (NTC) template. Both probes containing locked nucleic acid nucleotides were able to specifically amplify the template. NTC = no template control.
SNP analysis
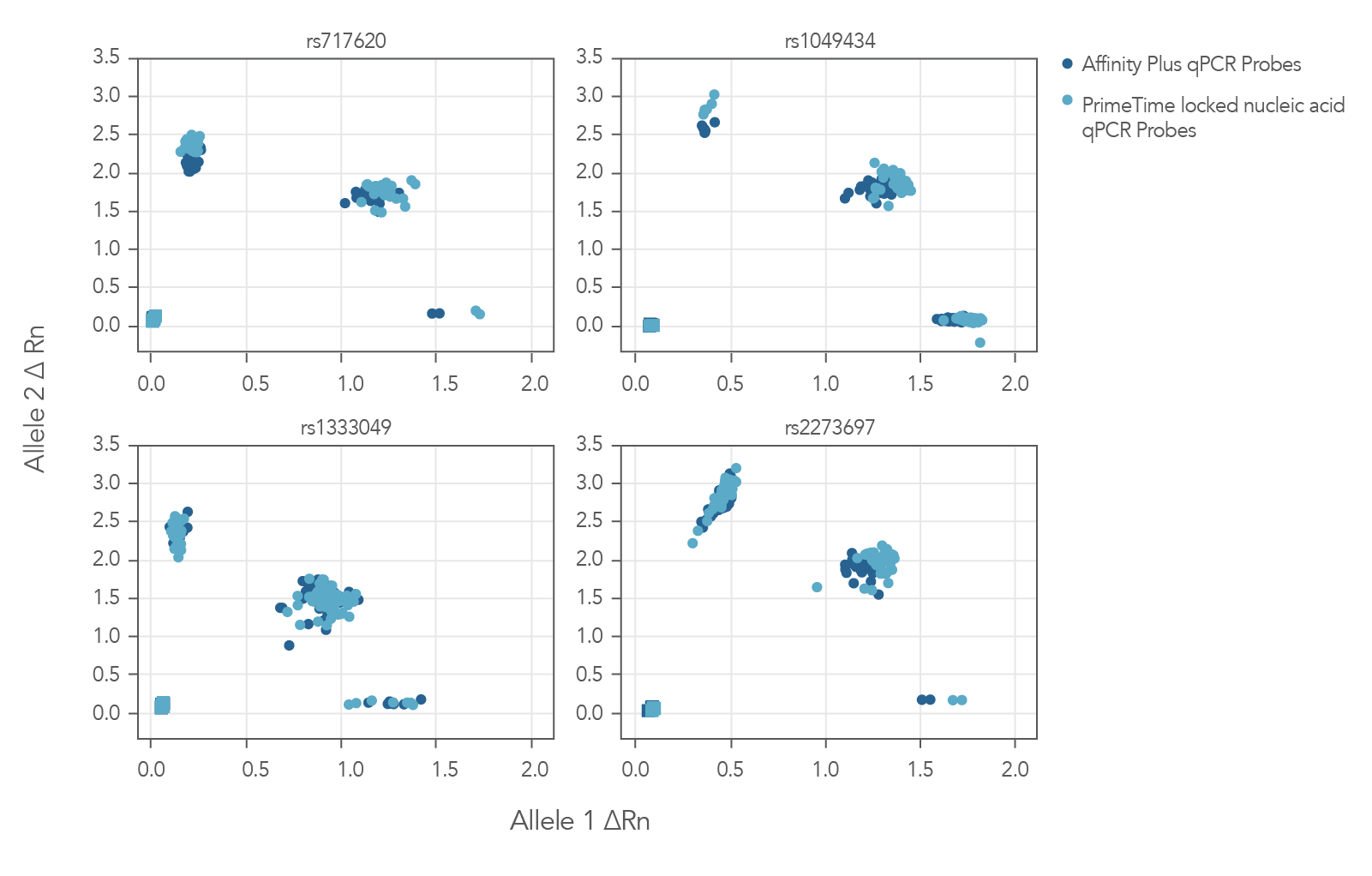
The Figure 3 cluster plots from 4 genotyping assays demonstrate that locked nucleic acid containing Affinity Plus qPCR Probes generate identical genotyping calls as PrimeTime locked nucleic acid probes. The probe sequences for both Affinity Plus and PrimeTime locked nucleic acid probes used in the assays are shown below in Table 1 (see Figure 3 sequences).
Figure 3. Locked nucleic acid probes synthesized as Affinity Plus qPCR Probes generate identical genotyping calls as PrimeTime locked nucleic acid probes. Genotyping assays (n = 4) were run using 46 unique Coriell gDNA samples and the same probe sequences synthesized as either Affinity Plus qPCR Probes or PrimeTime LNA qPCR Probes. There were no differences in calls observed between Affinity Plus and PrimeTime qPCR LNA probes. All genotype calls matched previous results (data not shown). The sequences of the probes are listed in Table 1 (see Figure 3 sequences). All but rs1333049 are ADME (absorption, distribution, metabolism, and excretion) assays.
Table 1. Probe, primer, and target sequences used to generate data presented in Figures 1−3.
| Name | Sequence (+N = Affinity Plus or locked nucleic acid nucleotide) |
|---|---|
| Figure 1 sequence | |
| Affinity Plus sequence | GGTCCT+T+A+CTTGGTG |
| Other locked nucleic acid sequence | GGTCCT+T+A+CTTGGTG |
| Figure 2 sequences | |
| M2-2 forward primer | GAGTCACTATGTATAATCAAAAAAACAC |
| M2-2 reverse primer | TGATCCGGTGGGTATATATGG |
| PrimeTime locked nucleic acid qPCR probes | /56-FAM/TA+T+CAA+T+CA+AAA+CAA+CC/3IABkFQ/ |
| Unmodified DNA | /56-FAM/TATCCATCAAAACAACC/3IABkFQ/ |
| M2-2 gBlocks Gene Fragment | TTGCGGAGGGAAGCTCATCAGTGGGGCCACGAGCTGAGTGCGTCCTGTCACTCCACTCCC ATGTCCCTTGGGAAGGTCTGAGACTAGGGTTAGAGTCACTATGTATAATCAAAAAAACACAC TATATATCAATCAAAACAACCAAAATAGCCATATATACCCACCGGATCAA |
| Figure 3 sequences | |
| rs717620_C | /56-FAM/AG+TCT+T+C+GTT+CCA/3IABkFQ/ |
| rs717620_T | /5YakYel/AGTCT+T+T+GTT+C+CAG/3IABkFQ/ |
| rs1049434_A | /56-FAM/CC+CTC+C+A+TCT+GTG/3IABkFQ/ |
| rs1049434_T | /5YakYel/C+CCTC+C+T+TCTGTGT/3IABkFQ/ |
| rs1333049_G | /56-FAM/A+C+AGT+T+G+AAAAGC/3IABkFQ/ |
| rs1333049_C | /5YakYel/AACAGT+T+C+AAA+A+GCA/3IABkFQ/ |
| rs2273697_C | /56-FAM/CT+CCA+A+C+GGT+GTAC/3IABkFQ/ |
| rs2273697_T | /5YakYel/TCTCCA+A+T+GGT+G+TAC/3IABkFQ/ |
Resources
Frequently asked questions
How do I dilute and store PrimeTime™ qPCR Probes?
We recommend resuspending PrimeTime qPCR Probes in TE (10 mM Tris pH 8.0, 0.1 mM EDTA). Alternatively, molecular grade water can be used. IDT recommends storing in a -20ºC freezer.
We find it convenient to initially make a 100 µM stock solution (which should be thawed relatively infrequently). Adding a volume of TE (µL) equal to 10 x # nmol DNA present in the tube (as noted on the spec sheet provided with the oligo) will produce a stock solution at this concentration.
Related products
References
1. Davalieva K, Kiprijanovska S, Plaseska-Karanfilska D. Fast, reliable and low cost user-developed protocol for detection, quantification and genotyping of hepatitis C virus. J Virol Methods. 2014;196:104-112.
2. Owczarzy R, You Y, Groth CL, et al. Stability and mismatch discrimination of locked nucleic acid-DNA duplexes. Biochemistry. 2011;50(43):9352-9367.
3. You Y, Moreira BG, Behlke MA, et al. Design of LNA probes that improve mismatch discrimination. Nucleic Acids Res. 2006;34(8):e60.
4. Johnson MP, Haupt LM, Griffiths LR. Locked nucleic acid (LNA) single nucleotide polymorphism (SNP) genotype analysis and validation using real-time PCR. Nucleic Acids Res. 2004;32(6):e55.