Whole exome sequencing
Tomorrow’s cancer research breakthroughs are on the horizon. xGen™ NGS for whole exome sequencing includes offerings for both routine and difficult samples—designed to minimize efforts while maintaining key data metrics and positioning your research for limitless potential.
xGen™ NGS—made limitless.
Overview
- Recommended products—curated for whole exome sequencing
- A tested solution—takes you from sample to sequencer
- Flexible sizing—available for 16 or 96 reactions
- Enzymatic fragmentation—no need for specialized equipment for library preparation
- Individually synthesized probes—quality-controlled probes, manufactured to ISO standards, with deep, uniform coverage
- Quality and consistency—high-quality metrics, displayed consistently across lots
- Automation friendly—easy workflow that can be automated for high-throughput applications
What is whole exome sequencing?
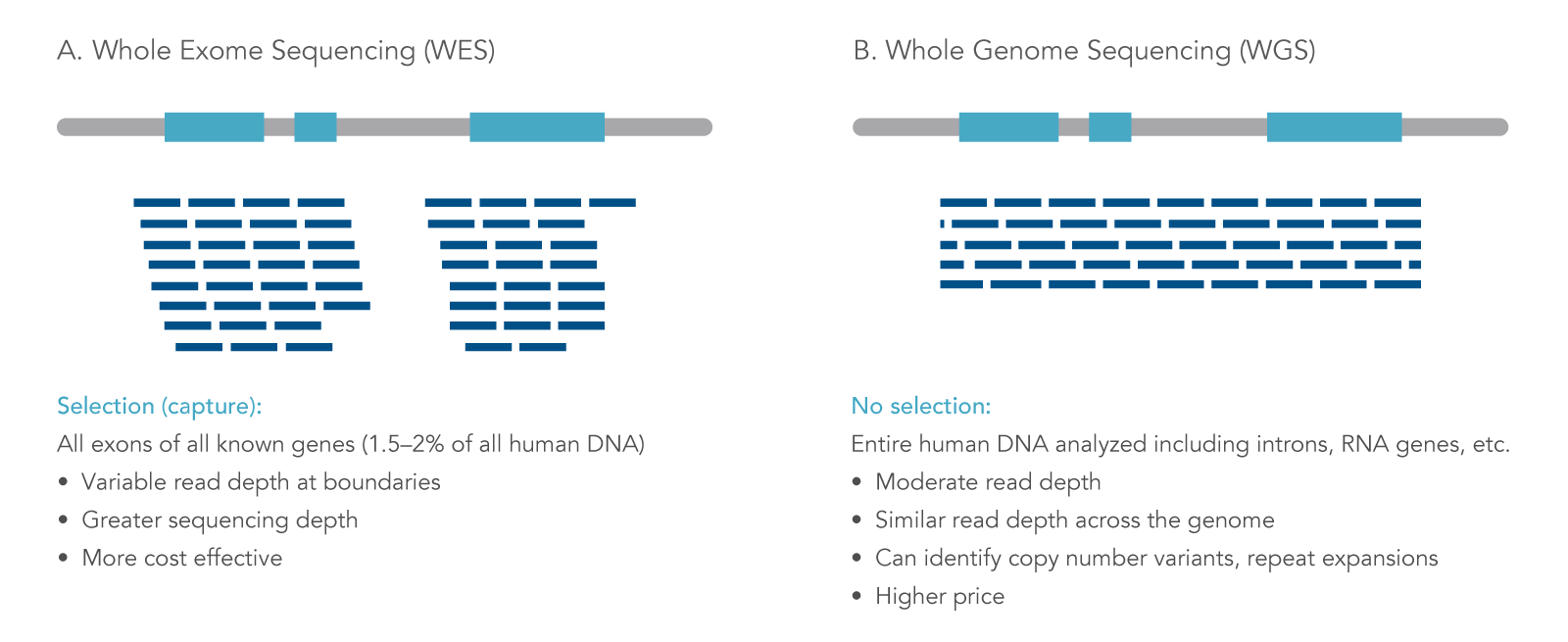
Whole exome sequencing (WES) is a targeted next generation sequencing (NGS) approach that uses modified oligonucleotide probes to “capture” and enrich the protein coding regions (exons) in a genome. Solely focusing on exons lowers the cost and time of sequencing as exons make up approximately 1% of the genome, but contain 85% of the variants associated with disease [1]. WES is a practical application for various fields of research such as population genetics and oncology research. Also, WES can be a feasible option for discovery science and data mining when searching for associations or linking genes to phenotypes [2]. The information gained from WES can provide researchers insights into germline and somatic variants within exons of genes. WES differs from other types of whole or targeted sequencing in several key ways (Figure 1).
Figure 1. Next generation sequencing approaches. NGS can be applied for different uses, including (A) whole exome sequencing (WES), where sequencing reads are focused on protein-coding regions of the genome; and (B) whole genome sequencing, where both coding and non-coding DNA regions are analyzed [3].
xGen whole exome sequencing solution
For WES, samples can be prepared using multiple library preparation methods. The xGen whole exome sequencing solution employs the xGen DNA Library Prep Kit EZ, which uses enzymatic fragmentation to shear high-quality DNA samples. This kit uses a TA ligation-based library prep and includes
an enzyme mix for fragmentation, end-repair, and A-tailing, as well as a ligase for the addition of the xGen Stubby Adapter (included in the xGen DNA Library Prep Kit EZ).
The xGen WES solution calls for premixed xGen UDI Primer Pairs for indexing by PCR, which generates NGS libraries that can be directly used for hybridization capture with the xGen Exome Hyb Panel v2. This panel consists of 415,115 individually synthesized, 5’ biotinylated, oligonucleotide probes. Each probe is quality controlled and combined equimolarly to help to provide ideal uniformity of enrichment from each panel. In an internal study, when compared to panels from other vendors, the xGen Exome Hyb Panel v2 had the highest on-target rate and mean coverage, targeted more exons with higher coverage depth, and had a more flexible workflow in terms of multiplexing range and hybridization times. Refer to the white paper, The xGen Exome Hyb Panel v2 – human exome sequencing with consistent coverage, for details on this comparison study.
The xGen whole exome sequencing solution includes xGen Hybridization Capture Core Reagents such as the xGen Hybridization and Wash v2 Kit, xGen Universal Blockers TS, and xGen Library Amplification Primer Mix. These reagents, when paired with the xGen Hyb Panel v2, help to provide a thorough, high-quality, whole exome sequencing workflow.
Extraction
LIBRARY PREP
xGen DNA Library Prep Kit EZ (16 or 96 rxn) for high-quality DNA samples
xGen UDI primers (16 or 96 rxn)
Sequencing & analysis
Method data
Library preparation and hybridization capture
To assess the xGen Exome Hyb Panel v2, twelve libraries were prepared using 100 ng of human genomic DNA (Coriell Institute). Then, xGen Stubby Adapters and Unique Dual Indexing primers (UDIs) were added with 6 cycles of PCR. The libraries were quantified using Qubit® dsDNA HS Assay Kit (Thermo Fisher Scientific). Indexed DNA libraries were captured and multiplexed according to the xGen Hyb and Wash Reagents v3 Kit protocol. Then, to test the flexibility of the xGen Exome Hyb Panel v2 in library capture strategies, 500 ng of a DNA library was used to generate four single-plex captures and one 8-plex capture (5 total captures). Each pool underwent a 4 h hybridization reaction. PCR was performed on each capture using settings selected for the panel size and the number of pooled libraries in each capture (10 PCR cycles for single-plex captures; 7 PCR cycles for 8-plex captures).
Whole Exome Sequencing
The final enriched library was normalized to 4 nM and sequenced on a NextSeq® 550 instrument (Illumina®) using 2 x 150 paired-end (PE) reads. The library was subsampled to 50 M total reads before analysis against the product’s target space in the human reference genome, hg38. Sequencing metrics were calculated using Picard analysis tools [4].
xGen DNA Library Prep EZ libraries in combination with the xGen Exome Hyb Panel v2 results in comprehensive and quality coverage
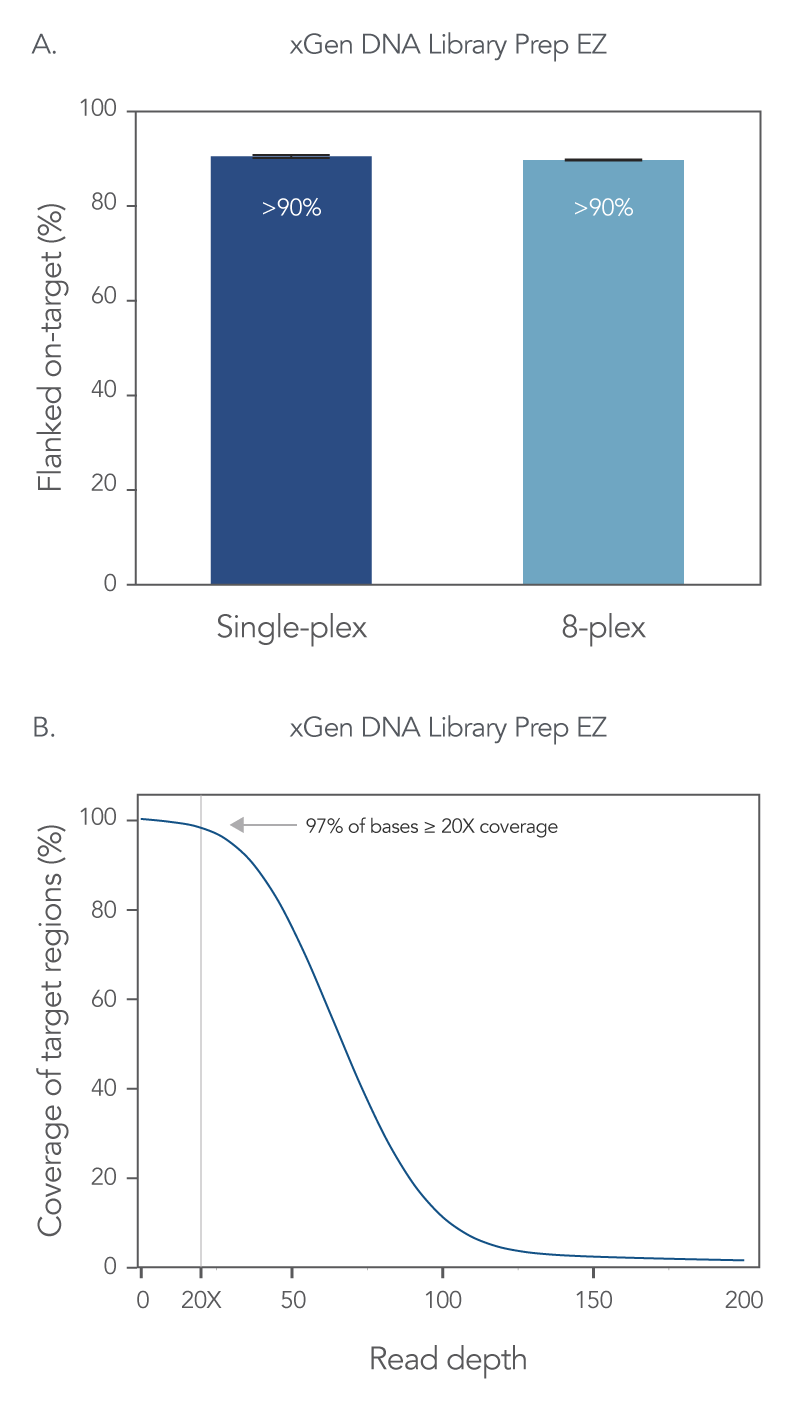
On-target rates give information regarding precision of bases or reads aligning with a targeted region, so a higher percentage of on-target reads indicates better capture precision. Using the xGen DNA Library Prep EZ kit and the xGen Exome Hyb Panel v2 an on-target rate of >90% was obtained for both single-plex and 8-plex captures (Figure 2A). In addition, with this WES workflow, 97% of the target bases reached >20X coverage depth with no noticeable drop-off in resolution with 8-plex captures (Figure 2B).
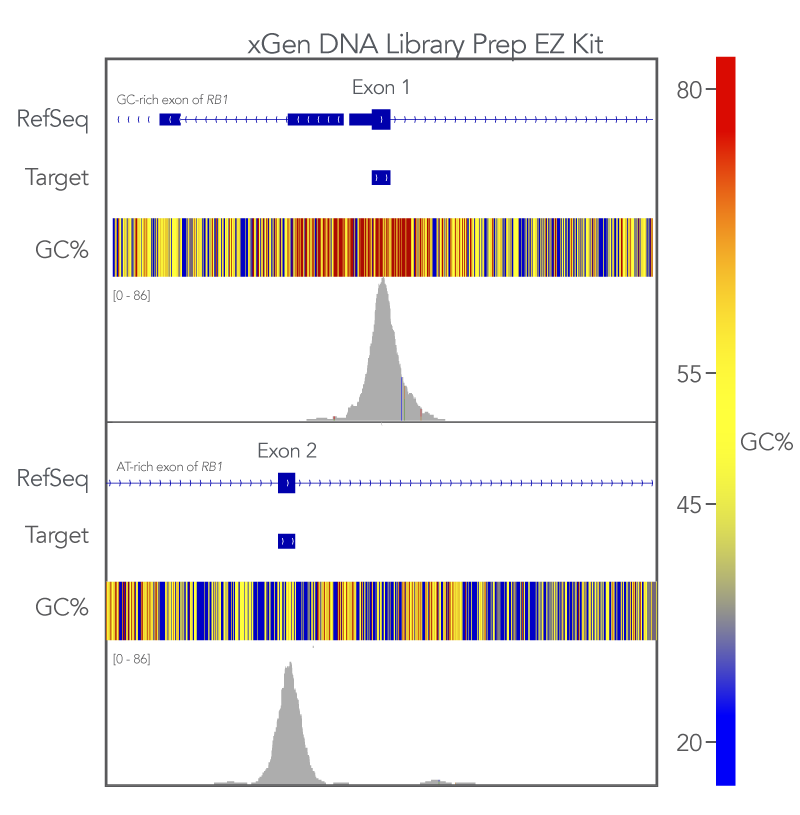
Uniform coverage of both high and low GC regions indicates more comprehensive coverage of targeted regions and fewer dropouts. To evaluate the xGen whole exome sequencing solution against this metric, Exon 1 (GC-rich) and Exon 2 (AT-rich) of the RB1 gene target space were analyzed. The sequences generated using the xGen Exome Hyb v2 panel uniformly covered both regions (Figure 3). Meaning that with the xGen whole exome sequencing solution, researchers get the uniform, comprehensive breadth of coverage as well as sufficient coverage depth that is necessary for reliable variant calling.
Figure 2. xGen whole exome sequencing solution offers comprehensive coverage and read depth. Libraries were prepared using 100 ng of human genomic DNA (Coriell Institute), and the xGen DNA Library Prep EZ Kit. Four single-plex WES captures, and one 8-plex WES capture were performed with the xGen Exome Hyb Panel v2. All captures used 500 ng of library and a 4 h hybridization reaction. The final library size was determined using a TapeStation® 4200 system and High Sensitivity D1000 ScreenTape® assay (Agilent); the final library concentration was determined using AccuClear® Ultra High Sensitivity dsDNA Quantitation kit (Biotium). Sequencing was completed on a NextSeq® 550 high-output flow cell (Illumina®) to generate 2 x 150 PE reads. Reads for all samples were subsampled to 50 M total reads. (A) Comparable flanked on-target rates are seen with both single-plex and 8-plex capture conditions. Error bars represent the standard deviation of replicates. (B) 97% of target bases are covered at ≥ 20X coverage. Post-capture libraries from these figures were amplified using KAPA 2x HiFi PCR Mix, as these data were generated prior to release of xGen 2x HiFi PCR Mix.
Figure 3. xGen DNA Library Prep EZ libraries in combination with the xGen Exome Hyb Panel v2 result in high-quality WES. (A) Integrated Genome Viewer (IGV) [5] plots show comprehensive coverage for both GC- and AT-rich regions of the RB1 gene. Post-capture libraries from these figures were amplified using KAPA 2x HiFi PCR Mix, as these data were generated prior to release of xGen 2x HiFi PCR Mix.
Ordering
Resources
References
- Choi M, Scholl UI, Ji W, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci U S A. 2009;106(45):19096-19101.
- Bamshad MJ, Ng SB, Bigham AW, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12(11):745-755.
- Gorcenco S, Ilinca A, Almasoudi W, et al. New generation genetic testing entering the clinic. Parkinsonism Relat Disord. 2020;73:72-84.
- “Picard Toolkit.” Broad Institute, GitHub repository: Broad Institute; 2019.
- Robinson JT, Thorvaldsdottir H, Winckler W, et al. Integrative Genomics Viewer. Nat Biotechnol. 2011;29(1):24-26.