Cas9 proteins
Experience the power of Alt-R Cas9 enzymes with widely adopted S. pyogenes Cas9, or our proprietary engineered Cas9 mutants with high editing potency and reduced off-target editing, or visualize and isolate edited cells with Alt-R Cas9-GFP/RFP Nucleases.
Ordering
- From standard (or wild type) to high-fidelity variants, Alt-R Cas9 enzymes provide unmatched accuracy and flexibility for diverse genome editing applications
- Reduce off-target editing with Alt-R HiFi Cas9 enzyme and explore specialized functions with nickases or dCas9 protein
- Streamline your workflow with GFP/RFP Cas9 fusion proteins for easy post-transfection tracking and cell sorting
Alt-R HiFi Cas9 Nuclease with improved selectivity
The new Alt-R HiFi Cas9 Nuclease has similar on-target potency to wild-type S.p. Cas9, but with significantly reduced off-target effects, allowing for precise genome editing, even under challenging conditions.
GMP CRISPR Nucleases
Does your project require high-quality GMP nucleases? SpCas9 Nuclease and SpyFi® Cas9 Nuclease are available for immediate delivery at GMP quality. Learn how IDT can help you rapidly transition from the lab to clinical trials. Contact us today.
Custom CRISPR solutions
Don’t see what you’re looking for? We are continually expanding our CRISPR product line, and we may have what you need. If you are interested in custom libraries, other CRISPR enzymes, formulations, or other CRISPR tools, contact our CRISPR experts today to discuss customized solutions for your research. Alt-R CRISPR enzymes are available in a variety of formats, with stock sizing available up to 50 mg. Larger formats and lot matching are also available upon request! Contact our experts.
Product Details
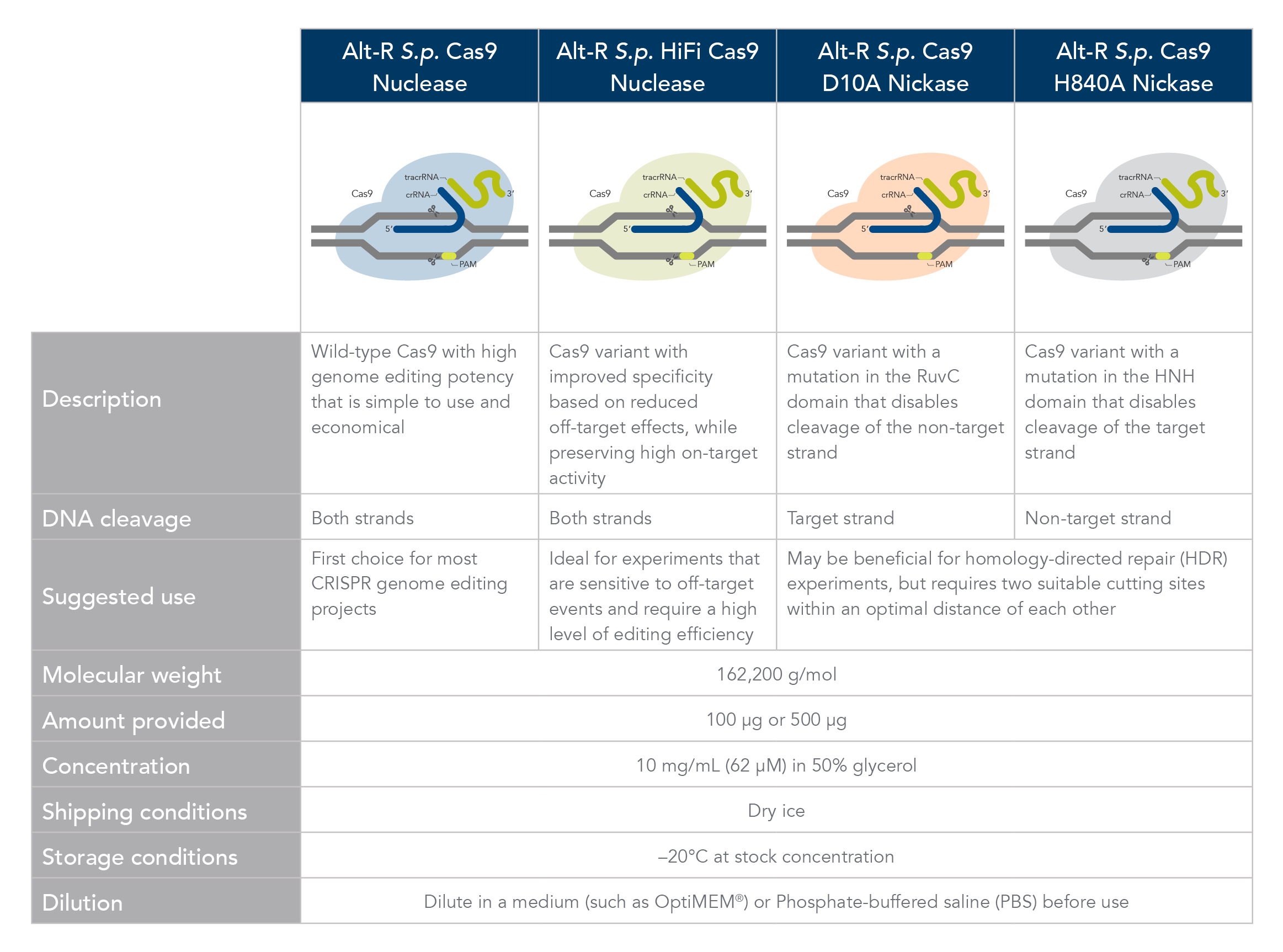
Which Cas9 enzyme to use
Comparison of Alt-R Cas9 Nucleases and Nickases. Click here to download PDF version.
Alt-R S.p. Cas9 nuclease
Alt-R S.p. Cas9 Nuclease V3 is the standard Cas9 used for general genome editing. It is a high purity, recombinant S. pyogenes Cas9. The enzymes include nuclear localization sequences (NLSs) and C-terminal 6-His tags. The S. pyogenes Cas9 enzyme must be combined with a gRNA to produce a functional, target-specific editing complex. For the best editing, combine the Alt-R S.p. Cas9 Nuclease V3 enzyme with Alt-R CRISPR gRNA in equimolar amounts.
Alt-R S.p. HiFi Cas9 nuclease
Alt-R S.p. HiFi Cas9 Nuclease V3 is also used for general genome editing, but it offers improved specificity over wild-type Cas9, greatly reducing the risk of off-target cutting events. This Cas9 variant also preserves the high level of editing efficiency expected from a Cas9 nuclease, maintaining 90–100% on-target editing activity at most sites. For applications that are sensitive to off-target events, combining the Alt-R S.p. HiFi Cas9 Nuclease V3 with the optimized Alt-R CRISPR‑Cas9 gRNA (crRNA:tracrRNA) is highly recommended.
Alt-R S.p. Cas9-GFP (or RFP) nuclease
The Alt-R S.p. Cas9-GFP V3 and S.p. Cas9-RFP V3 nucleases are high purity, recombinant S. pyogenes Cas9 enzymes that are expressed as fusion proteins with nuclear localization sequences (NLSs) and C-terminal 6-His tags. These enzymes have on-target functionality comparable to wild-type S.p. Cas9 and are designed for applications that require post‑transfection visualization of the protein or enrichment of edited cells using fluorescence-activated cell sorting (FACS). These enzymes should be combined with Alt-R CRISPR gRNA in equimolar amounts.
Alt-R S.p. Cas9 nickases
Cas9 nickases allow specific cutting of only one strand at the DNA target site. Cuts to both strands of DNA are accomplished by using either Alt-R S.p. Cas9 D10A Nickase V3 or Alt-R S.p. Cas9 H840A Nickase V3, with two gRNAs that target two neighboring Cas9 sites, one on either strand of the target region. There are two main reasons to consider using nickases. First, the use of two neighboring gRNAs instead of one gRNA (as used with Alt-R S.p. Cas9) can decrease off-target effects. Second, the rate of HDR is increased. For more information about using Cas9 nickases, see the application note.
Alt-R S.p. dCas9 protein
Alt-R S.p. dCas9 Protein V3 has mutations that result in the loss of nuclease activity. This protein can form RNP complexes with Alt-R gRNAs and bind to the target region specified by the gRNA without cutting the DNA. The primary use of dCas9 protein is to block transcription at a specific site on the genome. This is known as CRISPRi and is an alternative to RNAi for knockdown instead of knockout of genes.
Like the other Alt-R enzymes, Alt-R S.p. dCas9 Protein V3 is provided as 10 mg/mL in 50% glycerol, and it can be diluted in PBS or Opti-MEM media before use.
Product Data
Cas9 Nuclease
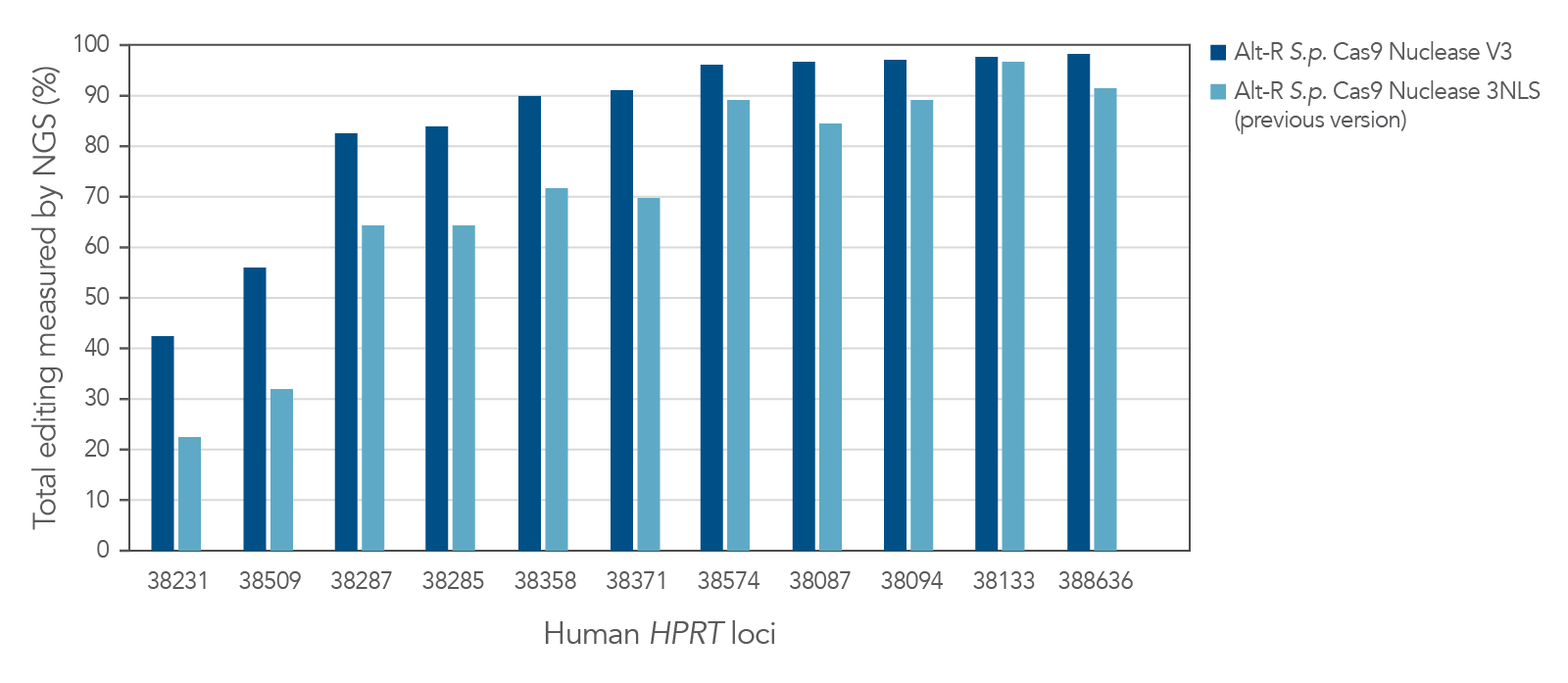
Improved editing efficiency using Alt-R S.p. Cas9 Nuclease V3
Alt-R S.p. Cas9 Nuclease V3 is designed to maximize the efficiency of genome editing across a broad number of sites. Modification of the expression construct facilitates nucleus-targeted delivery, resulting in enhanced cleavage, particularly at difficult targets (Figure 1).
Figure 1. Alt-R S.p. Cas9 Nuclease V3 genome editing efficiency even at challenging target sites. Ribonucleoprotein (RNP) complexes were formed with 1 of the 2 wild-type Cas9 proteins—Alt-R S.p. Cas9 Nuclease 3NLS (light blue) or Alt-R S.p. Cas9 Nuclease V3 (dark blue), combined with an Alt-R crRNA:tracrRNA complex targeting one of 11 loci on the human HPRT gene. RNP complexes (4 µM) were delivered into HEK-293 cells by nucleofection. Total editing at the on-target loci was calculated by NGS. n = 1.
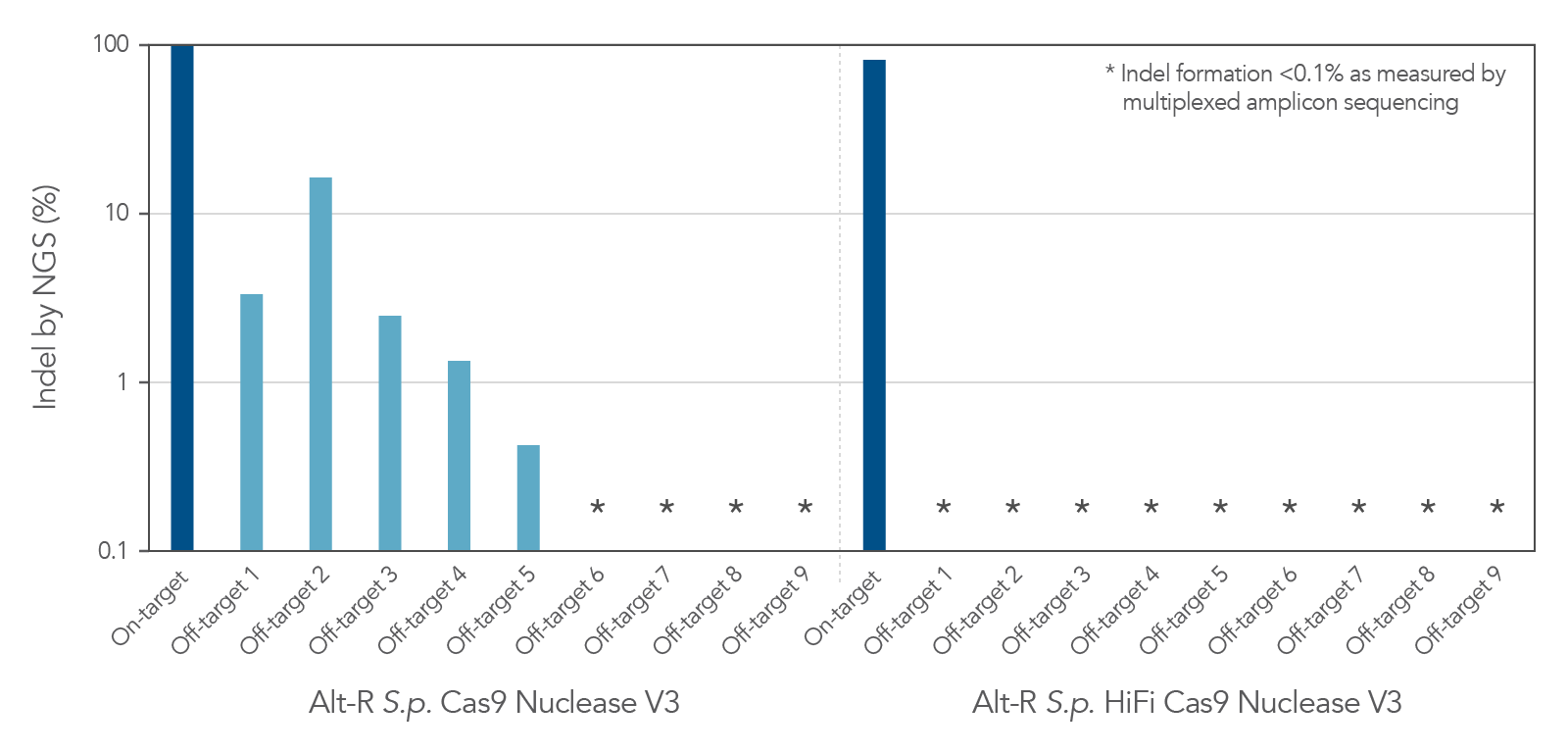
Increased specificity using Alt-R S.p. HiFi Cas9 Nuclease V3
As with the wild-type Alt-R Cas9 Nuclease V3, modification of the expression construct facilitates nucleus-targeted delivery, resulting in enhanced on-target cleavage by Alt-R S.p. HiFi Cas9 Nuclease V3. However, Alt-R HiFi Cas9 Nuclease V3 also provides superior cutting specificity (minimized off-target editing; Figure 2).
Figure 2. Alt-R S.p. HiFi Cas9 Nuclease V3 facilitates near-WT on‑target editing potency and reduces off-target site editing. RNP complexes were formed with either Alt-R S.p. Cas9 Nuclease V3 or Alt-R S.p. HiFi Cas9 Nuclease V3, combined with an Alt-R crRNA:tracrRNA complex targeting the EMX1 gene. RNP complexes (4 µM) were delivered into HEK-293 cells via nucleofection. Indel formation at the on-target locus as well as nine known off-target sites were measured by NGS (indicated on the y axis in log scale). n = 1.
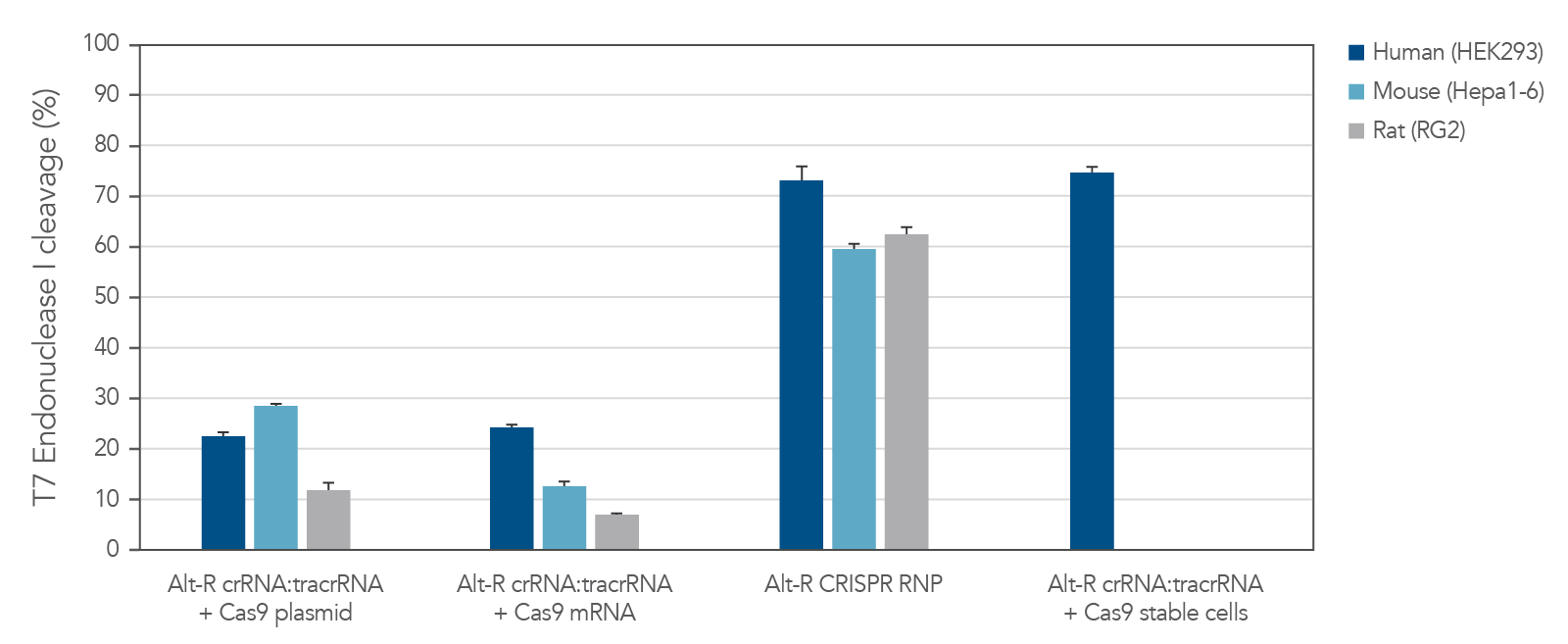
Potent editing with the Alt-R S.p. Cas9 nucleases
The Alt-R CRISPR-Cas9 System includes potent Alt-R S.p. Cas9 nucleases. When Alt-R S.p. Cas9 Nuclease 3NLS was combined with the Alt-R CRISPR crRNA and tracrRNA into a ribonucleoprotein (RNP), the system outperformed other editing approaches (Figure 3). You can expect even better editing efficiency with Alt-R S.p. Cas9 Nuclease V3 (see Figure 2). RNP transfections also provide control of amount of editing complexes used, and the non-renewable Cas9 RNP is cleared after a short duration by endogenous mechanisms, limiting off-target editing.
Figure 3. Experiment showing that lipofection of Alt-R CRISPR‑Cas9 System components as a ribonucleoprotein outperforms other transient CRISPR-Cas9 approaches. Alt-R CRISPR HPRT Control crRNA complexes for human, mouse, or rat were complexed with Alt-R CRISPR tracrRNA. Resulting complexes were transfected with Cas9 expression plasmid, Cas9 mRNA, or as part of a Cas9 RNP (containing Alt-R S.p. Cas9 Nuclease 3NLS, pre-complexed with the crRNA and tracrRNA) into human (HEK-293), mouse (Hepa1-6), or rat (RG2) cell lines. The Cas9 RNP outperformed the other transient Cas9 expression approaches, and performed similar to reference HEK293-Cas9 cells that stably express S. pyogenes Cas9. Error bars represent SD, n = 3.
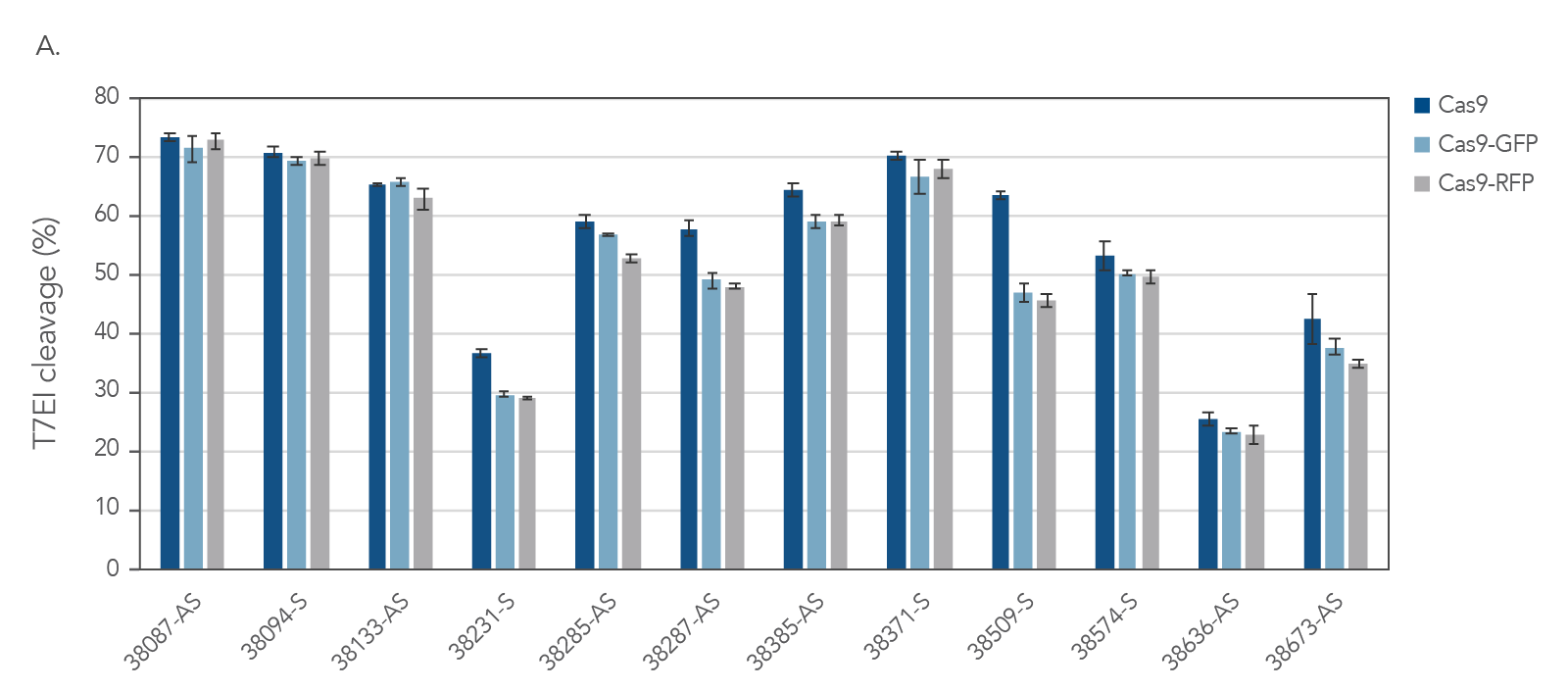
IDT’s Alt-R S.p. Cas9-GFP and Alt-R S.p. Cas9-RFP nucleases maintain consistent on‑target activity
Figure 4. IDT fluorescent CRISPR proteins maintain consistent on-target activity across multiple guides. Alt-R CRISPR-Cas9 sgRNAs were designed to target NGG PAM sites within the human HPRT gene. Guides were complexed with Alt-R S.p. Cas9 Nuclease V3, Cas9-GFP, or Cas9-RFP to form RNPs. RNPs were then delivered into HEK293 cells using the Lonza 96 well shuttle nucleofector at a concentration of 2.0 µM. After 48 hrs, genomic DNA was isolated (QuickExtract™ solution, Epicenter), and editing was assessed by T7El mismatch endonuclease assay. Error bars represent SD, n = 3.
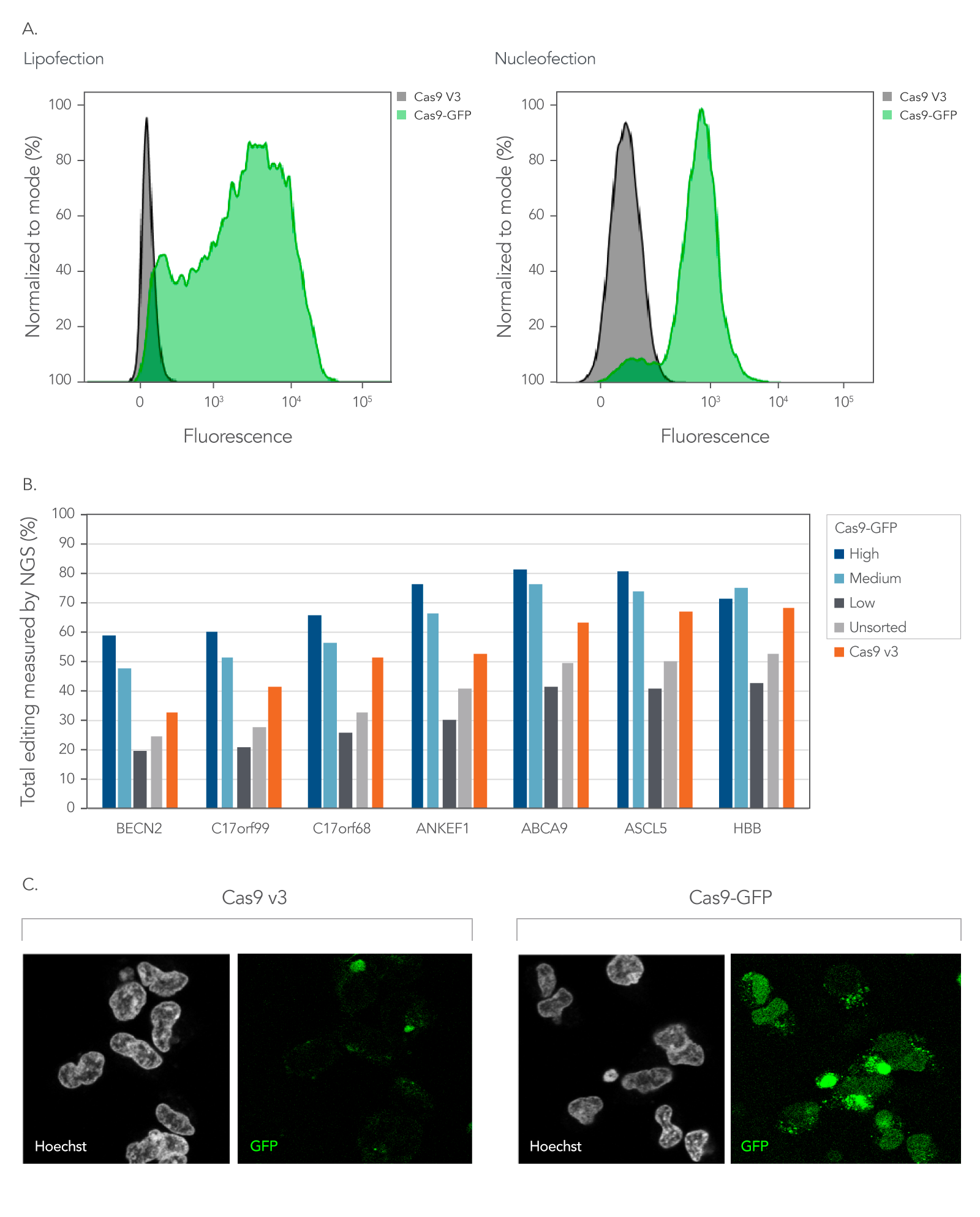
As shown in Figure 5, Alt-R fluorescent CRISPR nucleases enable enrichment of edited cells by fluorescent activated cell sorting (FACS).
Figure 5. Fluorescent CRISPR proteins can be used to enrich for edited cells by fluorescence activated cell sorting. (A) RNPs consisting of either Cas9-GFP or Cas9 V3 complexed with CRISPR‑Cas9 sgRNAs targeting two sites in the HPRT gene were delivered into HEK293 cells using either Lipofectamine™ RNAiMAX (Thermo Fisher) at 10 nM RNP or using a Nucleofector™ system (Lonza) at 2 µM RNP. The graphs show the GFP signal versus cell count, where cell count has been normalized to the mode for cells that had either Cas9-GFP or Cas9 V3 delivered using either lipofection or Nucleofection™. (B) Alt-R CRISPR-Cas9 sgRNAs were designed to target NGG PAM sites throughout the human genome. Guides were complexed with either Cas9-GFP or Alt-R Cas9 V3. RNPs were delivered into HEK293 cells using Lipofectamine™ RNAiMAX at 10 nM final concentration. After ~18 hrs, cells were sorted using a FACSAria™ II (Becton Dickinson) cell sorter into three subpopulations, GFP High: top 20%, Medium: 80–60%, and Low: Bottom 60% of cells based on GFP signal. Cells were then replated, and genomic DNA was isolated after 48-72 hrs. Editing was analyzed by NGS, n = 1. (C) Confocal images of HEK293 cells taken approximately 18 hours after delivery of either Cas9-GFP or Alt-R Cas9 V3 protein complexed with Alt-R CRISPR-Cas9 sgRNA delivered by Nucleofection™ at 2 µM RNP. Prior to imaging, live cells were incubated with Hoechst 33342 and washed with PBS. Cells were imaged in complete media in a chambered coverglass using a Leica SP8 confocal microscope.
Alt-R Cas9 nuclease performs well in human primary T-cells
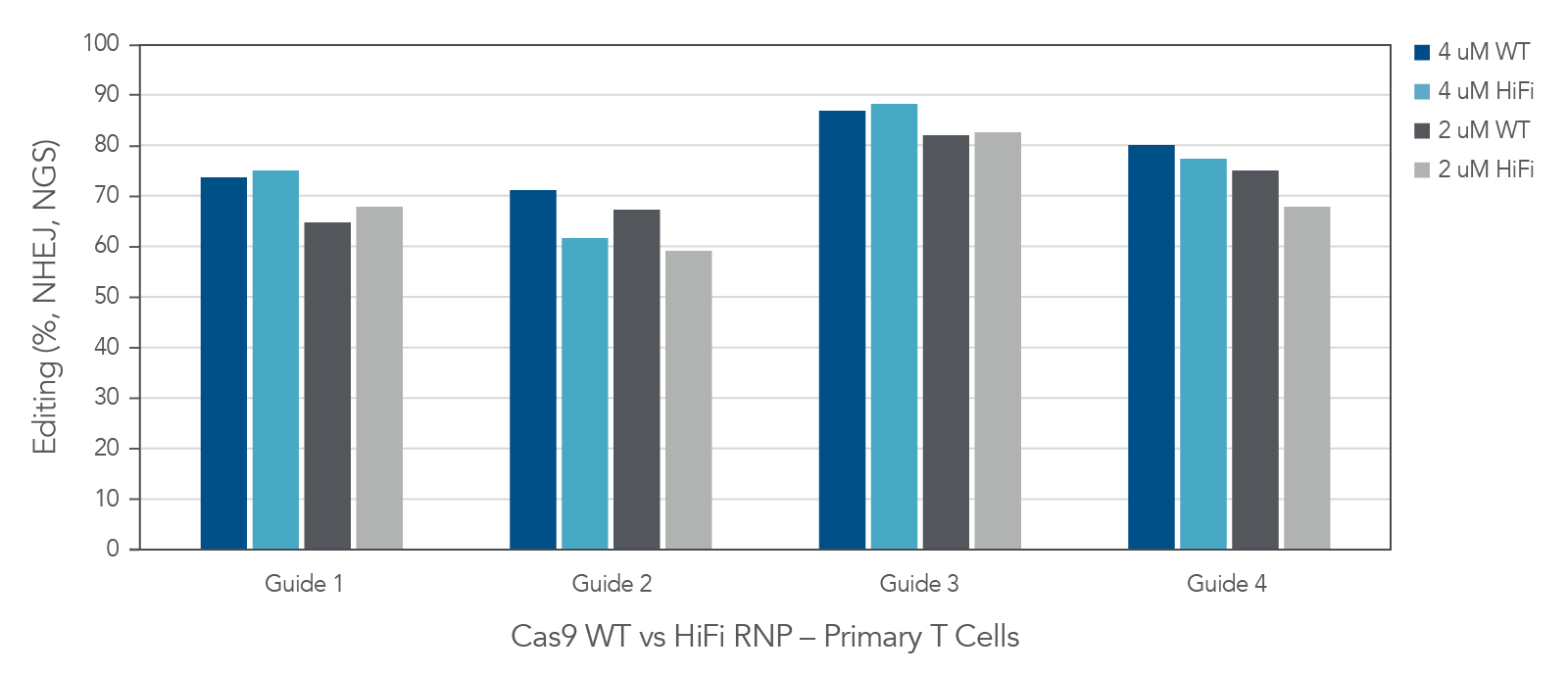
As the CRISPR/Cas9 system moves towards more therapeutic uses, it is important to observe how editing tools perform in cell types that are slightly more clinically relevant than easy-to-edit immortalized lines. Both our WT and HiFi Cas9 nucleases, when complexed with heavily modified Alt-R CRISPR sgRNA, are readily taken up by human primary T-cells and induce substantial levels of editing at different target sites (Figure 6).
Figure 6. In primary T-cells, wild type (Alt-R™ S.p. Cas9 Nuclease V3) and high-fidelity (Alt-R™ S.p. HiFi Cas9 Nuclease V3) Cas9 nucleases deliver high editing efficiency. Four highly-modified Alt-R™ CRISPR-Cas9 sgRNAs targeting individual genomic loci were complexed at a 1.2:1 sgRNA:Protein ratio with either Alt-R™ S.p. Cas9 Nuclease V3 or Alt-R™ S.p. HiFi Cas9 Nuclease V3. Following activation and a 48-hour incubation, single donor primary T-cells (Stemcell Technologies) were transfected with either 2 or 4 µM RNP alongside 4 µM Electroporation Enhancer using the Lonza 4D-Nucleofector® system. Following another 48-hour incubation period, genomic DNA was harvested and targeted amplicon sequencing was performed using the rhAmpSeq CRISPR Libraries Kit. Libraries were sequenced using the Illumina MiSeq platform, and editing rates were analyzed via an internal version of the rhAmpSeq CRISPR Analysis system. n = 1 transfection per condition.
*Small scale, standard, desalted oligos
GMP refers to manufacturing using a Quality Management System based on ISO 13485 standards and 21CFR820
Resources
Frequently asked questions
I transfected the cells with Alt-R™ S.p. Cas9-GFP. When should I sort the cells by fluorescence activated cell sorting (FACS) to enrich for transfected cells?
Our internal research data have shown that sorting cells within 24 hours after transfection of RNP complexes containing Alt-R S.p. Cas9-GFP nuclease gives detectable signal.
Can Alt-R™ S.p. Cas9-GFP be used with the same protocols as Alt-R S.p. Cas9 Nuclease V3?
Yes, Alt-R S.p. Cas9-GFP nuclease can be used in the same protocols and methods as the non-fluorescent Cas9 nucleases.
When should I consider using the glycerol-free Cas9 nuclease instead of the nucleases that include glycerol?
Alt-R™ S.p. Cas9 V3, glycerol-free may be of interest when working with samples or systems where the presence of glycerol may interfere, such as primary cell cultures or high-throughput instruments with sensitive fluidics.
When using the Alt-R™ S.p. HiFi Cas9 Nuclease V3, do I follow the same protocols for the standard Alt-R S.p. Cas9 Nuclease V3?
Yes. The Alt-R S.p. HiFi Cas9 Nuclease V3 can be directly substituted for the standard Alt-R S.p. Cas9 Nuclease V3 in the protocols.
When using the Alt-R™ Cas9 nickases, do I follow the same protocol as the standard Alt-R S.p. Cas9 Nuclease V3?
Yes. The delivery of the CRISPR-Cas9 ribonucleoprotein (RNP) complexes with Alt-R Cas9 nickases is similar to RNP with the standard Alt-R S.p. Cas9 Nuclease V3, so the same protocol can be used with these enzymes. However, with the Cas9 nickases, two guide RNAs are required to generate a double-strand break, while only one guide RNA is required for the standard nuclease.
When preparing materials to deliver two guide RNAs simultaneously, first prepare RNPs with each guide RNA separately before RNP delivery.
What is the on-target efficiency of the Alt-R™ S.p. HiFi Cas9 Nuclease V3?
The on-target efficiency ofAlt-R S.p. HiFi Cas9 Nuclease V3 is typically 90–100% of the standard Alt-R S.p. Cas9 Nuclease V3 efficiency at a specific target site.
The efficiency of both nucleases varies based upon the target site, cell type, and experimental conditions.
What is the expected reduction in off-target effects (OTEs), when using Alt-R™ S.p. HiFi Cas9 Nuclease V3?
In many cases, Alt-R S.p. HiFi Cas9 Nuclease V3 exhibits a 10–20 fold reduction in editing at problematic off-target sites, when compared with the standard Alt-R S.p. Cas9 Nuclease V3.
The amount of reduction in off-target editing is highly variable and can differ based on multiple factors, including off-target site, ribonucleoprotein (RNP) dose, and cell type.
What is the concentration of the Alt-R™ Cas9 nucleases and nickases?
The Alt-R Cas9 nucleases and nickases are supplied at concentration of 10 mg/mL in 25 mM Tris-Cl, 300 mM NaCl, 0.1 mM EDTA, 1 mM DTT, 50% glycerol (v/v), pH 7.4, with the exception of the Alt-R™ S.p. Cas9 V3, glycerol-free. The Alt-R™ S.p. Cas9 V3, glycerol-free is provided at a concentration of 10 mg/mL in a proprietary buffer without glycerol.
What buffer should I use to resuspend the Cas9 protein?
All Alt-R™ Cas9 nucleases and nickases are provided in solution at 10 mg/mL, so resuspension is not necessary.
Should I use the Alt-R™ Cas9 D10A or H840A nickase?
It depends upon the experimental design. Cas9 D10A will cut the target strand of DNA (which does not contain the PAM sequence), while Cas9 H840A will cut the non-target strand (which contains the PAM sequence). The availability and location of PAM sites within the target region of DNA needs to be assessed.
How should I store the Cas9 protein?
See our "Stability of CRISPR reagents under different freezing, storage, and thawing conditions" DECODED™ online article for more details.
How does Alt-R™ S.p. HiFi Cas9 Nuclease V3 differ from the standard Alt-R S.p. Cas9 Nuclease V3?
Alt-R S.p. HiFi Cas9 Nuclease V3 was developed to reduce off-target cutting events, with similar high, on-target activity as the Alt-R S.p. Cas9 Nuclease V3.
Alt-R HiFi Cas9 nuclease can be substituted for Alt-R Cas9 nuclease in experiments that are sensitive to off-target cutting events. We recommend testing a dose-response curve using Alt-R HiFi Cas9 or other Cas9 proteins to determine the optimal dosage.
https://www.idtdna.com/pages/products/crispr-genome-editing/alt-r-crispr-enzymes