Alt-R™ HDR Donor Templates
Enhanced UltramerTM DNA oligos specifically built for homology-directed repair (HDR) in research applications.
Our proprietary synthesis process delivers HDR-ready oligos of high quality up to 200 bases manufactured under ISO 9001 standards. For longer HDR donors, you can order our Alt-R HDR Donor Blocks. Alt-R HDR Donor Oligos are available in tube or plate formats.
Ordering
- Single-stranded DNA oligos built for homology-directed repair (HDR)
- Ideal for introducing point mutations or short insertions
- Optimized through extensive laboratory testing
HDR Positive Controls
Use the HDR positive controls below along with the Alt-R HDR Donor Oligos to help determine the HDR efficiency in your experiments. HDR positive controls provide different Cas9 gRNA and scale options and include tracrRNA along with human or mouse donor oligos. See the product sheet for additional information; research protocol details about the HDR positive controls are available in the Resources tab below.
| Guide RNA | Human (Homo sapiens) | Mouse (Mus musculus) |
|---|---|---|
| Alt-R™ CRISPR-Cas9 crRNA | Add to Cart | Add to Cart |
| Alt-R™ CRISPR-Cas9 crRNA XT | Add to Cart | Add to Cart |
| Alt-R™ CRISPR-Cas9 sgRNA | Add to Cart | Add to Cart |
Alt-R HDR Design Tool
If you don’t have a template design of your own, use our Alt-R HDR Design Tool to design your template. Simply provide basic information about your target site and then use the tool to design and visualize your desired edit within the sequence. The Alt-R HDR Design Tool will provide the recommended HDR donor template along with gRNA(s) for your specifications.
Alt-R™ HDR Enhancer Protein
Alt-R HDR Enhancer Protein is a novel protein-based reagent that promotes homology-directed repair by inhibiting 53BP1, a key regulator of dsDNA break repair pathway choice. It improves editing efficiency by up to 2X across established and hard-to-edit primary cells (iPSCs, HSPCs) without increasing off-target effects, genomic translocations, or compromising cell viability. Compatible with multiple CRISPR systems, including Cas9, Cas12a, and Eureca™-V, it integrates seamlessly into existing workflows. Offered in RUO format with a cGMP grade coming soon, Alt-R HDR Enhancer Protein is specifically engineered for translational researchers advancing CRISPR-based therapeutics.
Alt-R HDR Enhancer V2
Alt-R HDR Enhancer V2 is a small molecule compound that increases homology-directed repair by blocking the NHEJ pathway. Alt-R HDR Enhancer V2 exhibits its activity in multiple cell lines, including both adherent and suspension cell lines. Its activity is independent of the enzyme employed; for example, it can be used either with Alt-R S.p. Cas9 nucleases or A.s. Cas12a (Cpf1) nucleases. This versatile reagent is also compatible with electroporation and lipofection methods. When used in combination with Alt-R HDR Donor Oligos, Alt-R HDR Enhancer V2 significantly improves knock-in efficiency, increasing precise HDR events. See below data for details.
Request a consultation
Have questions for our experts? Your time is valuable and we’ll prioritize your inquiry. Click on “Request a consultation” to provide brief information about your project, and we’ll be in touch to discuss it ASAP.
Request a consultationProduct Details
Homology-directed repair (HDR) donors
IDT provides different HDR donor options for researchers to select from, depending on the specific needs of their application:
- No modification—standard DNA
- PS modification—4 phosphorothioate bonds: 5′–N*N*NNN…NNN*N*N–3′, where * represents a PS bond and N represents a DNA base
- Alt-R HDR modification—IDT’s proprietary modification pattern developed through extensive testing to increase donor oligo stability and enable the highest rate of repair
Product Data
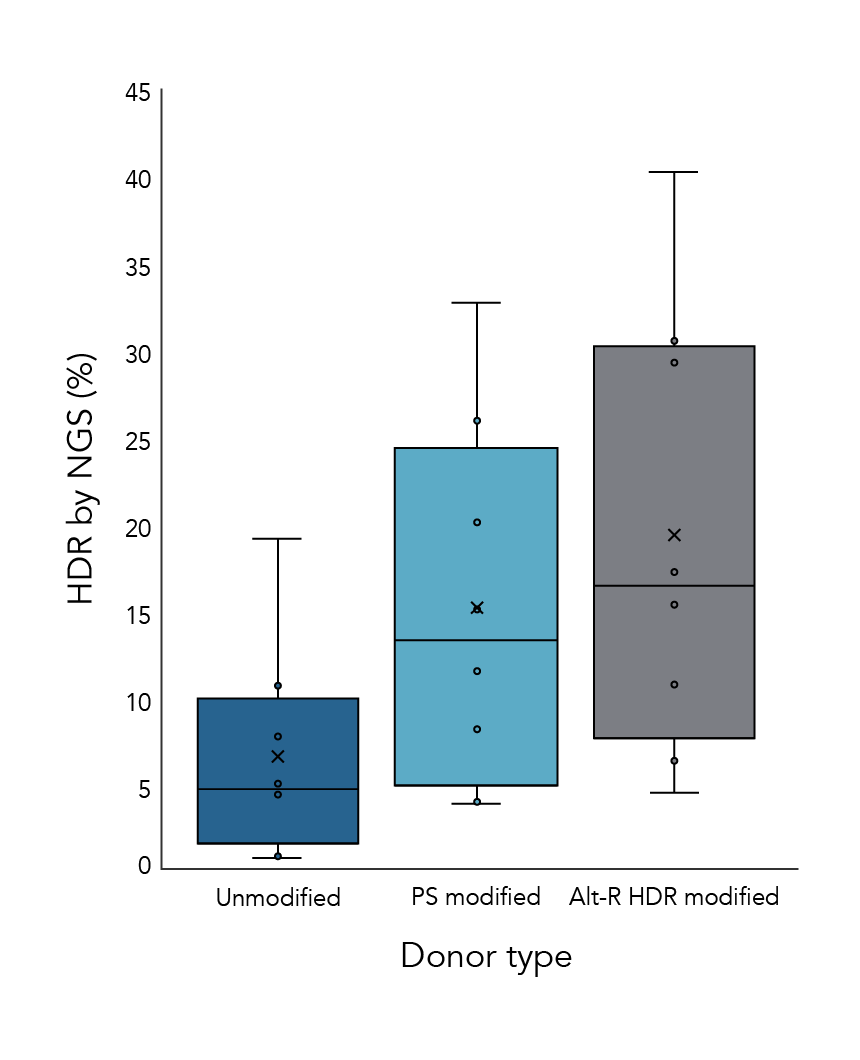
Alt-R HDR Donor Oligos improve HDR efficiency
The HDR rates from experiments using 3 different donor options were compared. Donor oligos with Alt-R HDR modifications showed increased HDR rates compared to other formats (Figure 1).
Figure 1. Alt-R HDR donors improve HDR efficiency over other donor types. HeLa and Jurkat cells were electroporated with 2 µM Cas9 RNP complexes (Alt-R S.p. HiFi Cas9 Nuclease V3 complexed with Alt-R CRISPR-Cas9 crRNA and tracrRNA) targeting 4 genomic loci along with 0.5 µM single-stranded HDR donor template using the Nucleofection™ System (Lonza). Donor templates contained no modifications (Unmodified), 4 phosphorothioate linkages (2 at each end of the template; PS modified), or the Alt-R HDR modification (Alt-R HDR modified). Genomic DNA was isolated 48 hours (HeLa) or 72 hours (Jurkat) after electroporation, and HDR efficiency was measured by amplicon sequencing on the MiSeq system (Illumina, v2 chemistry, 150 bp paired-end reads).
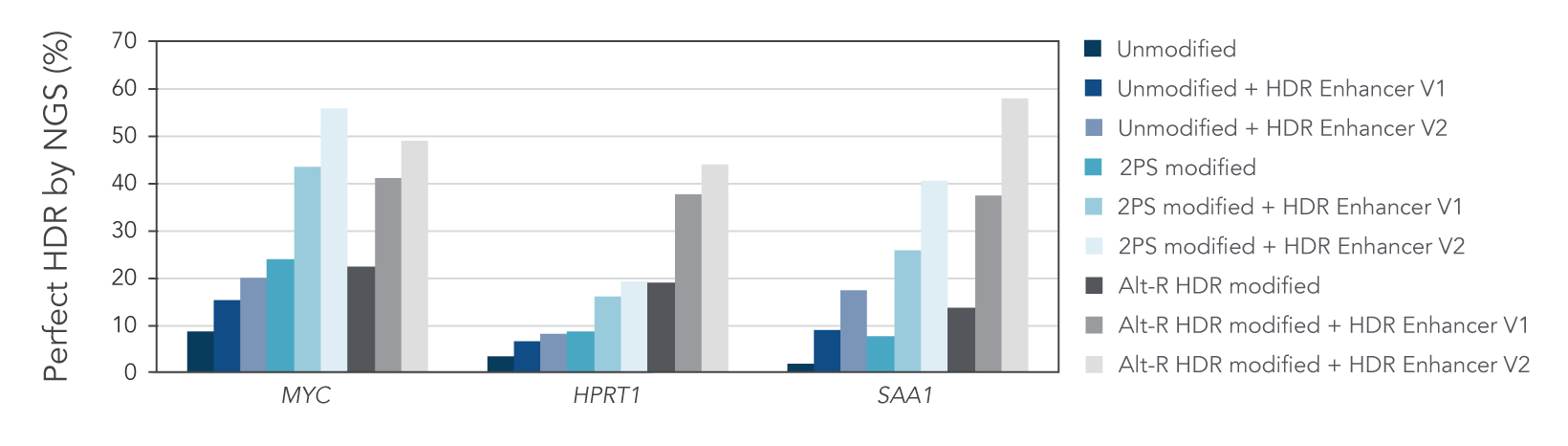
Alt-R HDR modified donors and Alt-R HDR Enhancer lead to higher HDR rates
We investigated whether combining Alt-R HDR Enhancer with Alt-R modifications of HDR donor oligos would improve HDR rates further. Our data demonstrated that this works well, leading to higher HDR rates (Figure 2).
Figure 2. Alt-R HDR modified donors and Alt-R HDR Enhancer have an additive effect on HDR improvement. RNP complexes (2 μM) targeting three genomic loci (MYC, HPRT1, and SAA1) along with 0.5 μM single-stranded Alt-R HDR donor oligo were delivered to HeLa cells by electroporation with 3 μM Alt-R Cas9 Electroporation Enhancer using the 4D-Nucleofector™ System (Lonza). The RNP complex comprised Alt-R S.p. HiFi Cas9 Nuclease V3 complexed with Alt-R CRISPR-Cas9 crRNA and tracrRNA. Unmodified, PS modified, or Alt‑R HDR modified donor templates were used. Immediately after electroporation, cells were plated in media with either no treatment, 30 μM Alt-R HDR Enhancer V1, or 1 μM HDR Enhancer V2. Genomic DNA was isolated 48 hours after electroporation, and HDR efficiency was measured by amplicon sequencing on the MiSeq system (Illumina, v2 chemistry, 150 bp paired-end reads). The highest HDR rate was achieved with the combination of the Alt-R HDR Donor and HDR Enhancer V2.
Resources
Frequently Asked Questions
CRISPR genome editing efficiency seems low - any steps to improve this?
In addition, not every sequence associated with a PAM site performs the same. For example, polymorphisms in the protospacer binding site may reduce editing efficiency. Base mismatches also become more detrimental to editing the closer they are to the PAM site.
We recommend that you try 2 or 3 different PAM sites in your gene of interest to identify a site that provides optimal editing efficiency. Also, be sure to include control experiments. Alt-R™ CRISPR-Cas9 HPRT Positive Controls and Alt-R™ CRISPR-Cas9 Negative Controls are available for human, mouse, and rat.
Please feel free to contact us if you need additional assistance; having the results of your control experiments available will facilitate our ability to help you. For more information, go to www.idtdna.com/ContactUs.
When should I consider using the glycerol-free Cas9 nuclease instead of the nucleases that include glycerol?
Alt-R™ S.p. Cas9 V3, glycerol-free may be of interest when working with samples or systems where the presence of glycerol may interfere, such as primary cell cultures or high-throughput instruments with sensitive fluidics.
Do you offer CRISPR gRNA libraries?
Yes, we offer predesigned Cas9 CRISPR crRNA and additional options for creating customizable gRNA libraries.
For more information, please visit our Custom guide RNA Libraries page.
Can I use a CRISPR-Cas9 crRNA protospacer sequence that is shorter than 20 nt?
The Alt-R™ CRISPR-Cas9 crRNA ordering tool (accessible here) accommodates 19 and 20 nucleotide (nt) protospacer sequences; however, we recommend 20 nt sequences for most experiments. Other formats can be ordered as custom RNAs.
There are reports in the literature suggesting that CRISPR-Cas9 nuclease specificity can be improved through use of truncated guide RNAs [1]. For example, 17 nt protospacer elements have been reported to reduce off-target effects.
In contrast, our research investigating the effect of shorter protospacer element length on CRISPR-Cas9 nuclease specificity demonstrated that 20 nt protospacer elements were optimal, with 19 nt protospacers providing similar strong editing efficacy in most cases. When using Alt-R S.p. HiFi Cas9 Nuclease, 20 nt protospacer sequences provide the greatest amount of genomic editing.
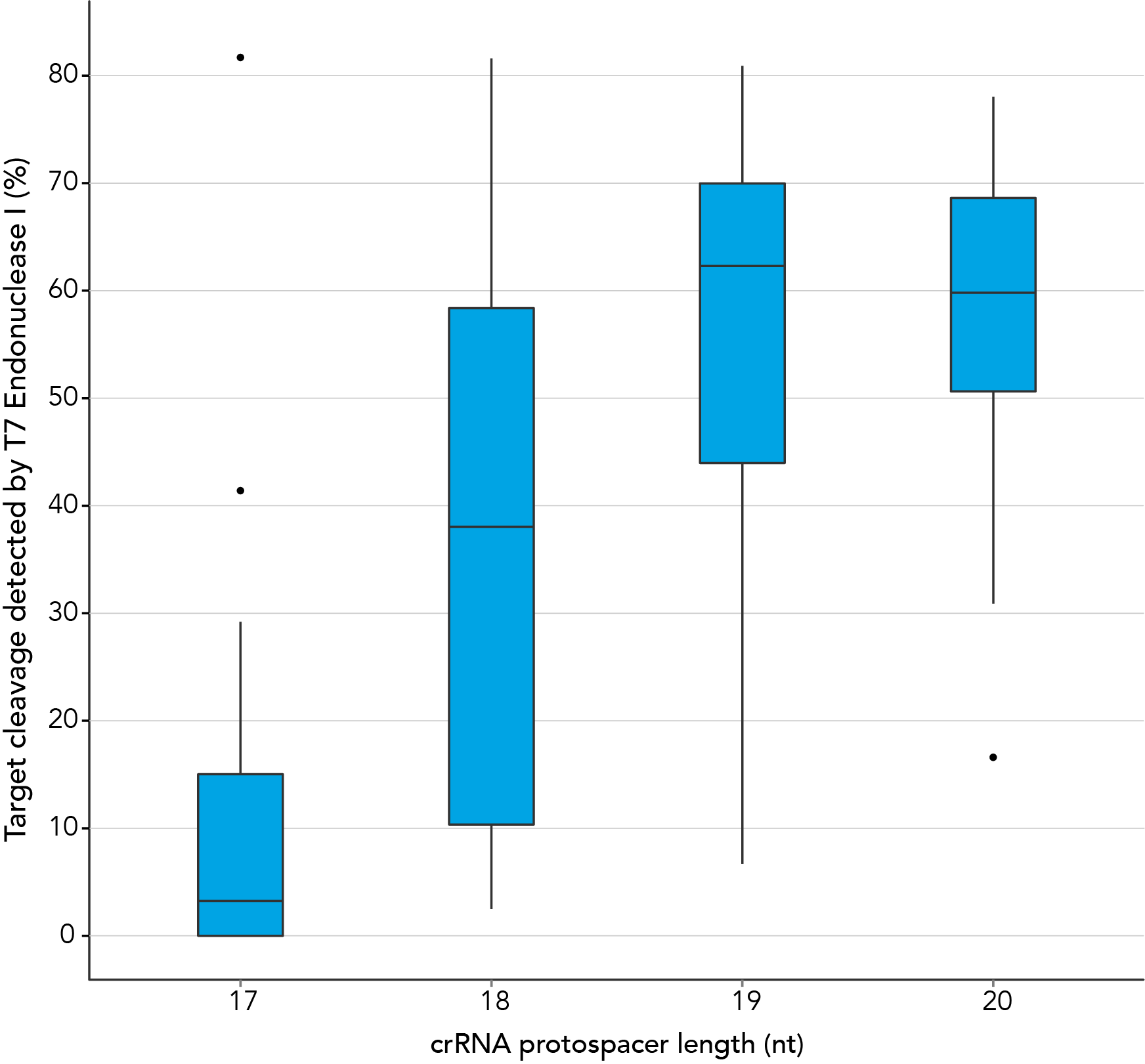 Figure 1. 19-20 nt protospacer element provides optimal genome editing. crRNAs with varying protospacer element lengths (17–20 nt) were designed to 12 distinct HPRT target sites. crRNA:tracrRNA complexes were reverse transfected into a HEK-293 cell line stably expressing S. pyogenes Cas9, using Lipofectamine® RNAiMAX Transfection Reagent (Thermo Fisher Scientific). Genomic DNA was isolated and editing was measured by PCR amplification of target sites followed by cleavage with T7EI mismatch endonuclease (Alt-R™ Genome Editing Detection Kit) and analysis using the Fragment Analyzer™ (Advanced Analytical). At all but one of the 12 target sites, crRNAs with 19 and 20 base protospacer elements produced the greatest amount of genomic editing. Each of the 12 data points for each category represent the average of 3 biological replicates, with the exception of one data point in the 19 base category that is composed of 2 biological replicates.
Figure 1. 19-20 nt protospacer element provides optimal genome editing. crRNAs with varying protospacer element lengths (17–20 nt) were designed to 12 distinct HPRT target sites. crRNA:tracrRNA complexes were reverse transfected into a HEK-293 cell line stably expressing S. pyogenes Cas9, using Lipofectamine® RNAiMAX Transfection Reagent (Thermo Fisher Scientific). Genomic DNA was isolated and editing was measured by PCR amplification of target sites followed by cleavage with T7EI mismatch endonuclease (Alt-R™ Genome Editing Detection Kit) and analysis using the Fragment Analyzer™ (Advanced Analytical). At all but one of the 12 target sites, crRNAs with 19 and 20 base protospacer elements produced the greatest amount of genomic editing. Each of the 12 data points for each category represent the average of 3 biological replicates, with the exception of one data point in the 19 base category that is composed of 2 biological replicates.- Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32(3):279-284.
RUO21-0224_001.1