What is the value of assessing HRD in solid tumor samples?
Homologous recombination deficiency (HRD) is a genomic signature which indicates a dysfunction in homologous recombination repair (HRR). HRR plays a crucial role in maintaining genomic stability by facilitating the accurate repair of DNA double-strand breaks; when this pathway is impaired, it leads to genomic instability, making cells susceptible to accumulating mutations and increasing the risk of cancer development.
The identification of loss-of-function mutations in HRR genes such as BRCA1 and BRCA2 is one well established approach for determining HRD status, however this approach alone can miss some HRD-positive tumors. Other approaches include characterizing the changes that occur in a genome as a result of HRD causing genomic instability—i.e., genomic signatures or genomic scars such as loss of heterozygosity (LOH) and other large chromosomal re-arrangements. A composite HRD score combines information about multiple HRD genomic scars into a single numerical value.
Methods that rely on information from both gene-mutation approaches and genomic scar-based approaches help ensure that HRD-positive samples are correctly identified and allow labs to accurately stratify samples based on HRD status. This stratification is a key predictor as to whether a tumor would respond to certain molecular agents like PARP inhibitors, which have been demonstrated to be an effective lethal agent in HRD-positive tumors in ovarian, breast, and prostate cancers[1-3]. (Figure 1).
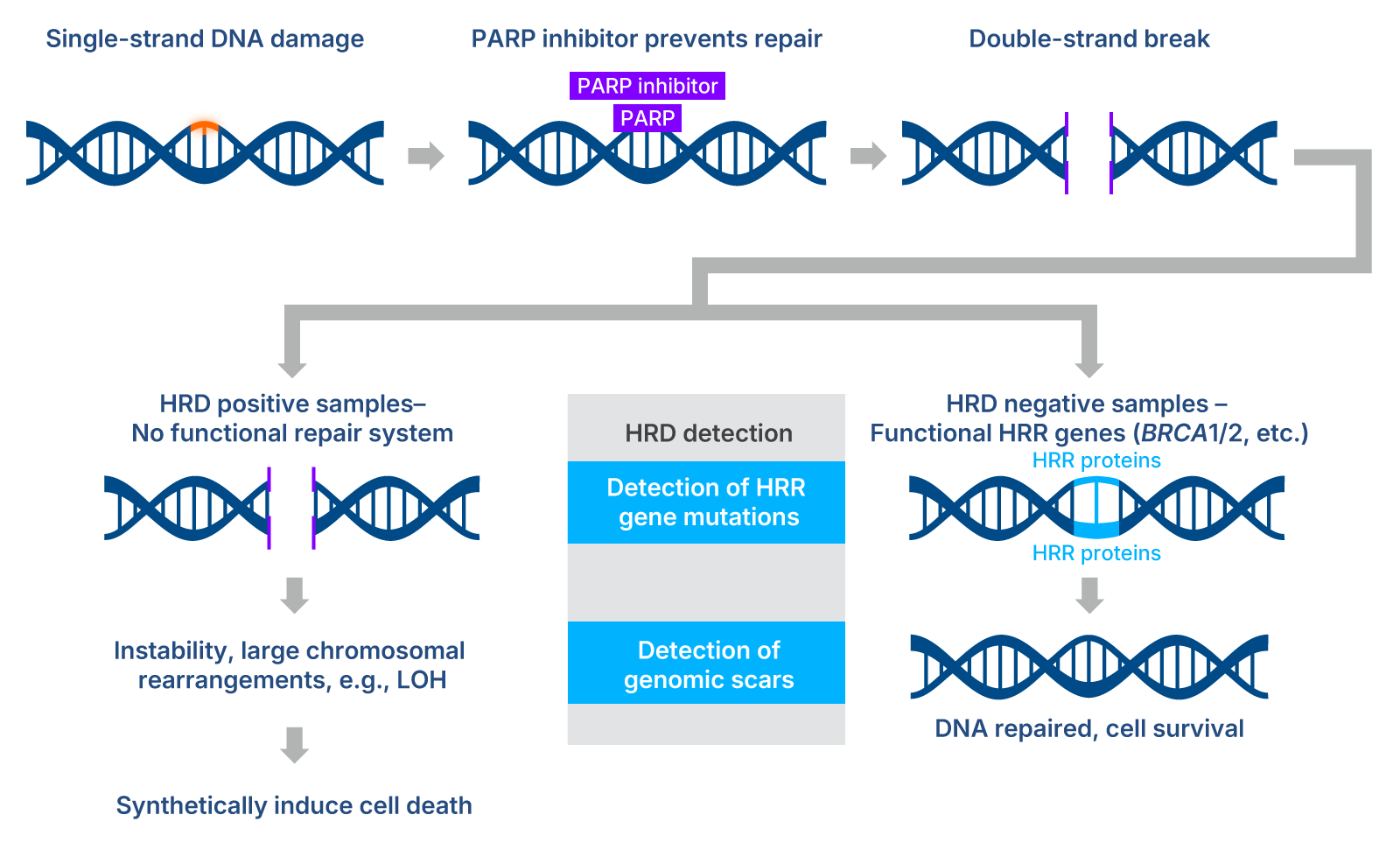
Figure 1. PARP inhibition of single-strand DNA repair results in targeted synthetic lethality in cells with homologous recombination deficiency. HRD detection approaches are largely based on targeting mutated HRR genes and/or identifying genomic scars like LOH. Adapted from[4].
What are the key features of a reliable HRD assessment?
High-quality, scalable HRD assessment workflows are:
- Comprehensive
- Reliable
- Flexible, modular, and automatable
Comprehensive: A truly comprehensive HRD assessment enables the detection of HRD-related alterations across different tumor types and provides insights into both HRR-related genes such as BRCA1, BRCA2, and PLAB2 as well as genomic instability signatures (e.g., LOH) in a single workflow. Additionally, a reliable and integrated bioinformatic analysis pipeline that identifies mutations and can correctly classify samples based on key genomic signatures is also a key component of a comprehensive HRD assessment.
For a more complete view of the molecular landscape of a tumor sample, approaches like comprehensive genomic profiling (CGP) provide insights into various genomic signatures like tumor mutational burden (TMB) and microsatellite instability (MSI), as well as HRD. Read more about the role of genomic signatures in sample stratification in the Comprehensive and streamlined genomic signature assessment white paper.
Reliable: A common approach to test the reliability of an assessment approach is the analysis of concordance data, which compares the classification of samples (e.g., HRD positive or negative) between different methodologies. When different results are obtained and non-concordant samples are identified, additional investigation is recommended to determine the true status of a sample.
An example of concordance data can be seen in Figure 2, where two composite HRD scoring methods are compared. Below the results from a VARIANTPlex Complete Solid Tumor v2 panel, which incorporates the assessment genomic scars like LOH is compared to another composite HRD score. These approaches were 92.3% concordant, suggesting that the VARIANTPlex Complete Solid Tumor v2 panel can reliability classify the HRD status of samples based on this high concordance to an orthogonal method.
VARIANTPlex solid tumor assays use AMP chemistry and Archer Analysis software (v7.4 or higher) to assess HRD. This involves analyzing allele-specific copy numbers and the percentage of indel variants related to non-homologous end joining (%NHEJ) to generate an HRD score. Samples are classified as positive, negative, or optional intermediate, with adjustable thresholds for flexibility.
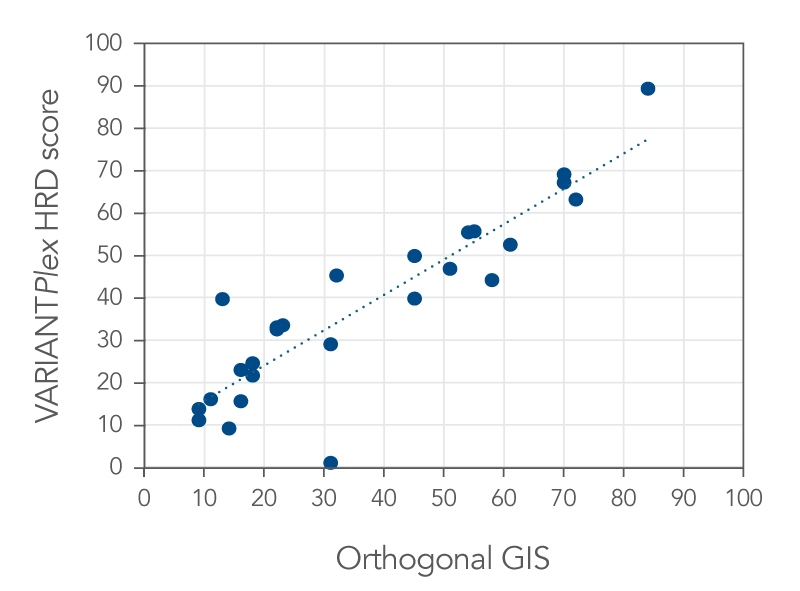
Figure 2. VARIANTPlex HRD assessment is highly concordant with orthogonal genomic instability score (GIS). Twenty-six samples were assessed with VARIANTPlex Complete Solid Tumor v2 and Archer Analysis 7.4. FFPE samples from solid tumors including ovarian, breast, cervical, lymph nodes and other sources (n = 22), a blood sample, and FFPE reference materials (n = 3). The reference materials included an HRD negative, an HRD low positive, and an HRD high positive sample, which were 100% concordant.
Flexible, modular, and automatable: As new relevant targets are identified, biomarker assays should be flexible enough to add content for those targets quickly and easily without impacting existing workflows. Modular HRD assessment gives labs that flexibility, saves important sequencing resources, and avoids sample depletion with the potential to assess samples that need only HRD classification. Automatable workflows can save hands-on time and minimize the risk of pipetting errors, giving lab personnel more time to complete additional high-priority tasks.
Access to flexible software analysis (i.e., secondary and tertiary analysis) that accurately and quickly provide labs with the answers they need is a key factor to consider when selecting an HRD assessment workflow. Analysis pipelines that allow labs to adjust key features, like the threshold to differentiate the HRD status of a sample, provides greater control and flexibility and offers the chance to adapt analyses to incorporate the latest findings in the field.
Labs can build a high-quality HRD assessment panel with Archer NGS solutions by designing a custom VARIANTPlex panel that includes insights into keys HRR genes (e.g., BRCA1, BRCA2, RAD51C, RAD51D, and PALB2) as well as an HRD composite score; or by integrating the VARIANTPlex BRCA v3 panel with the VARIANTPlex HRD Module. Download this flyer to learn more about Archer’s flexible HRD solutions; or request a consultation to learn more about Archer CGP assays with option HRD assessment.
Future of HRD in precision cancer research
While HRD assessment technologies have rapidly developed in recent years, key challenges remain to ensure that samples are reliably classified. First, labs need assessment approaches that provide them with the insights they need from limited samples with constrained resources (e.g., lab personnel and sequencing limitations). Additionally, harmonization of HRD assessment methods, addressing tumor heterogeneity, and understanding the functional consequences of different HRD alterations are ongoing areas of research. Finally, exploring novel biomarkers and refining computational approaches for HRD signature analysis will continue to enhance the accuracy of HRD assessments providing greater confidence to labs.

